Model info
| Transcription factor | GATA2 (GeneCards) | ||||||||
| Model | GATA2_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 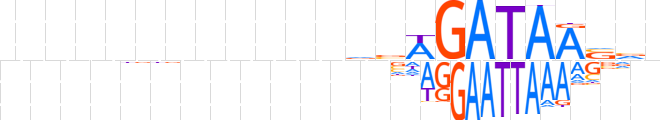 | ||||||||
| LOGO (reverse complement) | 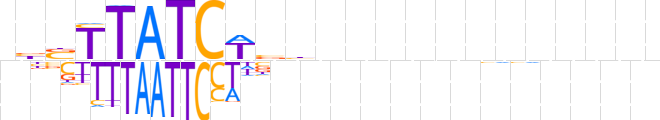 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnnnnnnnnnnnvvWGATAASvn | ||||||||
| Best auROC (human) | 0.976 | ||||||||
| Best auROC (mouse) | 0.978 | ||||||||
| Peak sets in benchmark (human) | 84 | ||||||||
| Peak sets in benchmark (mouse) | 54 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | GATA-type zinc fingers {2.2.1} | ||||||||
| TF subfamily | Two zinc-finger GATA factors {2.2.1.1} | ||||||||
| HGNC | HGNC:4171 | ||||||||
| EntrezGene | GeneID:2624 (SSTAR profile) | ||||||||
| UniProt ID | GATA2_HUMAN | ||||||||
| UniProt AC | P23769 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | GATA2 expression | ||||||||
| ReMap ChIP-seq dataset list | GATA2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 44.406 | 22.669 | 73.704 | 10.15 | 71.793 | 23.522 | 3.827 | 41.303 | 21.717 | 40.815 | 38.131 | 28.241 | 11.443 | 10.921 | 42.309 | 15.049 |
| 02 | 43.757 | 21.26 | 69.196 | 15.146 | 48.786 | 17.799 | 9.071 | 22.27 | 41.154 | 50.216 | 43.955 | 22.646 | 7.626 | 24.291 | 43.769 | 19.058 |
| 03 | 21.306 | 22.182 | 75.977 | 21.858 | 34.395 | 33.241 | 7.249 | 38.681 | 63.959 | 51.121 | 26.607 | 24.304 | 5.239 | 23.136 | 29.527 | 21.218 |
| 04 | 19.806 | 15.836 | 43.453 | 45.804 | 28.701 | 21.851 | 8.061 | 71.066 | 68.977 | 14.665 | 27.76 | 27.957 | 11.58 | 17.268 | 60.925 | 16.288 |
| 05 | 15.714 | 25.47 | 35.896 | 51.985 | 11.798 | 8.885 | 4.272 | 44.666 | 34.695 | 35.253 | 27.462 | 42.79 | 14.116 | 26.241 | 104.231 | 16.528 |
| 06 | 16.195 | 11.139 | 30.866 | 18.123 | 34.262 | 27.836 | 6.704 | 27.046 | 35.041 | 50.434 | 47.729 | 38.657 | 13.959 | 16.91 | 107.798 | 17.301 |
| 07 | 13.89 | 16.642 | 50.997 | 17.927 | 44.25 | 24.632 | 7.655 | 29.781 | 49.52 | 39.182 | 65.425 | 38.97 | 9.11 | 15.823 | 54.907 | 21.289 |
| 08 | 17.27 | 19.293 | 48.672 | 31.535 | 35.602 | 29.625 | 5.821 | 25.232 | 51.083 | 54.383 | 42.111 | 31.407 | 22.021 | 13.674 | 50.468 | 21.804 |
| 09 | 15.818 | 29.835 | 51.718 | 28.604 | 28.123 | 34.907 | 2.917 | 51.027 | 38.156 | 31.802 | 50.861 | 26.253 | 12.341 | 34.721 | 52.492 | 10.424 |
| 10 | 20.996 | 20.261 | 41.765 | 11.417 | 35.921 | 48.463 | 11.7 | 35.182 | 51.258 | 29.13 | 47.561 | 30.039 | 12.86 | 36.271 | 43.894 | 23.283 |
| 11 | 27.569 | 31.421 | 43.136 | 18.908 | 38.751 | 30.318 | 9.262 | 55.795 | 24.274 | 33.795 | 52.98 | 33.87 | 10.552 | 20.03 | 40.112 | 29.228 |
| 12 | 14.342 | 14.24 | 57.725 | 14.838 | 64.536 | 21.599 | 9.189 | 20.24 | 39.612 | 22.305 | 58.467 | 25.106 | 30.366 | 25.392 | 72.143 | 9.899 |
| 13 | 16.994 | 55.88 | 65.758 | 10.225 | 24.846 | 35.118 | 7.8 | 15.771 | 30.386 | 79.087 | 66.275 | 21.778 | 5.296 | 31.656 | 21.956 | 11.174 |
| 14 | 62.09 | 0.955 | 2.847 | 11.629 | 95.505 | 4.825 | 2.754 | 98.657 | 120.702 | 7.34 | 0.984 | 32.763 | 40.581 | 1.096 | 1.195 | 16.075 |
| 15 | 0.0 | 0.923 | 317.057 | 0.899 | 0.0 | 0.0 | 14.217 | 0.0 | 0.0 | 0.0 | 7.779 | 0.0 | 0.0 | 0.0 | 159.125 | 0.0 |
| 16 | 0.0 | 0.0 | 0.0 | 0.0 | 0.923 | 0.0 | 0.0 | 0.0 | 496.274 | 0.0 | 1.025 | 0.88 | 0.899 | 0.0 | 0.0 | 0.0 |
| 17 | 0.0 | 7.056 | 5.256 | 485.784 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.025 | 0.0 | 0.0 | 0.0 | 0.88 |
| 18 | 0.0 | 0.0 | 0.0 | 0.0 | 6.127 | 0.0 | 0.0 | 0.929 | 5.256 | 0.0 | 0.0 | 0.0 | 476.552 | 0.88 | 1.94 | 8.316 |
| 19 | 411.551 | 5.562 | 54.053 | 16.769 | 0.0 | 0.0 | 0.0 | 0.88 | 1.94 | 0.0 | 0.0 | 0.0 | 9.245 | 0.0 | 0.0 | 0.0 |
| 20 | 48.588 | 113.82 | 235.45 | 24.878 | 2.703 | 0.911 | 0.988 | 0.96 | 4.902 | 22.552 | 23.605 | 2.993 | 0.0 | 1.175 | 14.606 | 1.867 |
| 21 | 16.555 | 8.654 | 29.901 | 1.082 | 88.726 | 26.003 | 12.89 | 10.84 | 108.532 | 48.366 | 97.797 | 19.954 | 5.01 | 6.846 | 16.975 | 1.867 |
| 22 | 61.149 | 29.96 | 67.45 | 60.264 | 52.022 | 16.341 | 4.895 | 16.612 | 54.416 | 41.94 | 44.037 | 17.172 | 8.008 | 10.076 | 13.82 | 1.84 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.348 | -0.316 | 0.851 | -1.099 | 0.825 | -0.28 | -2.016 | 0.276 | -0.359 | 0.264 | 0.197 | -0.1 | -0.984 | -1.029 | 0.3 | -0.718 |
| 02 | 0.333 | -0.379 | 0.788 | -0.711 | 0.441 | -0.554 | -1.207 | -0.334 | 0.272 | 0.47 | 0.338 | -0.317 | -1.373 | -0.248 | 0.333 | -0.487 |
| 03 | -0.377 | -0.338 | 0.881 | -0.352 | 0.095 | 0.061 | -1.421 | 0.211 | 0.71 | 0.487 | -0.159 | -0.248 | -1.727 | -0.296 | -0.056 | -0.381 |
| 04 | -0.449 | -0.668 | 0.326 | 0.378 | -0.084 | -0.352 | -1.32 | 0.815 | 0.785 | -0.743 | -0.117 | -0.11 | -0.972 | -0.583 | 0.662 | -0.64 |
| 05 | -0.675 | -0.202 | 0.137 | 0.504 | -0.954 | -1.227 | -1.915 | 0.353 | 0.103 | 0.119 | -0.128 | 0.311 | -0.78 | -0.172 | 1.196 | -0.626 |
| 06 | -0.646 | -1.01 | -0.012 | -0.536 | 0.091 | -0.114 | -1.495 | -0.143 | 0.113 | 0.474 | 0.419 | 0.21 | -0.791 | -0.604 | 1.229 | -0.581 |
| 07 | -0.796 | -0.619 | 0.485 | -0.547 | 0.344 | -0.235 | -1.37 | -0.048 | 0.456 | 0.224 | 0.732 | 0.218 | -1.203 | -0.669 | 0.558 | -0.378 |
| 08 | -0.583 | -0.475 | 0.439 | 0.009 | 0.129 | -0.053 | -1.628 | -0.211 | 0.487 | 0.549 | 0.295 | 0.005 | -0.345 | -0.811 | 0.475 | -0.355 |
| 09 | -0.669 | -0.046 | 0.499 | -0.087 | -0.104 | 0.109 | -2.259 | 0.486 | 0.197 | 0.017 | 0.482 | -0.172 | -0.91 | 0.104 | 0.514 | -1.074 |
| 10 | -0.392 | -0.427 | 0.287 | -0.986 | 0.138 | 0.434 | -0.962 | 0.117 | 0.49 | -0.069 | 0.416 | -0.039 | -0.871 | 0.147 | 0.336 | -0.29 |
| 11 | -0.124 | 0.005 | 0.319 | -0.494 | 0.213 | -0.03 | -1.187 | 0.574 | -0.249 | 0.077 | 0.523 | 0.08 | -1.062 | -0.438 | 0.247 | -0.066 |
| 12 | -0.764 | -0.771 | 0.608 | -0.731 | 0.719 | -0.364 | -1.195 | -0.428 | 0.235 | -0.332 | 0.621 | -0.216 | -0.028 | -0.205 | 0.83 | -1.123 |
| 13 | -0.599 | 0.576 | 0.737 | -1.092 | -0.226 | 0.115 | -1.352 | -0.672 | -0.028 | 0.921 | 0.745 | -0.356 | -1.717 | 0.013 | -0.348 | -1.007 |
| 14 | 0.68 | -3.159 | -2.28 | -0.968 | 1.109 | -1.803 | -2.31 | 1.141 | 1.342 | -1.409 | -3.138 | 0.047 | 0.258 | -3.059 | -2.995 | -0.653 |
| 15 | -4.4 | -3.183 | 2.306 | -3.202 | -4.4 | -4.4 | -0.773 | -4.4 | -4.4 | -4.4 | -1.354 | -4.4 | -4.4 | -4.4 | 1.618 | -4.4 |
| 16 | -4.4 | -4.4 | -4.4 | -4.4 | -3.183 | -4.4 | -4.4 | -4.4 | 2.754 | -4.4 | -3.109 | -3.217 | -3.202 | -4.4 | -4.4 | -4.4 |
| 17 | -4.4 | -1.447 | -1.724 | 2.732 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.109 | -4.4 | -4.4 | -4.4 | -3.217 |
| 18 | -4.4 | -4.4 | -4.4 | -4.4 | -1.58 | -4.4 | -4.4 | -3.179 | -1.724 | -4.4 | -4.4 | -4.4 | 2.713 | -3.217 | -2.609 | -1.291 |
| 19 | 2.567 | -1.671 | 0.543 | -0.612 | -4.4 | -4.4 | -4.4 | -3.217 | -2.609 | -4.4 | -4.4 | -4.4 | -1.189 | -4.4 | -4.4 | -4.4 |
| 20 | 0.437 | 1.284 | 2.009 | -0.225 | -2.326 | -3.192 | -3.135 | -3.155 | -1.788 | -0.321 | -0.277 | -2.236 | -4.4 | -3.007 | -0.747 | -2.641 |
| 21 | -0.624 | -1.252 | -0.044 | -3.069 | 1.036 | -0.181 | -0.868 | -1.036 | 1.236 | 0.432 | 1.132 | -0.442 | -1.768 | -1.476 | -0.6 | -2.641 |
| 22 | 0.665 | -0.042 | 0.763 | 0.651 | 0.505 | -0.637 | -1.79 | -0.621 | 0.549 | 0.291 | 0.339 | -0.589 | -1.327 | -1.106 | -0.801 | -2.653 |