Model info
| Transcription factor | GATA2 (GeneCards) | ||||||||
| Model | GATA2_HUMAN.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 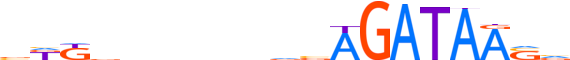 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bKKbnnnnndvAGATAASv | ||||||||
| Best auROC (human) | 0.958 | ||||||||
| Best auROC (mouse) | 0.971 | ||||||||
| Peak sets in benchmark (human) | 84 | ||||||||
| Peak sets in benchmark (mouse) | 54 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | GATA-type zinc fingers {2.2.1} | ||||||||
| TF subfamily | Two zinc-finger GATA factors {2.2.1.1} | ||||||||
| HGNC | HGNC:4171 | ||||||||
| EntrezGene | GeneID:2624 (SSTAR profile) | ||||||||
| UniProt ID | GATA2_HUMAN | ||||||||
| UniProt AC | P23769 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | GATA2 expression | ||||||||
| ReMap ChIP-seq dataset list | GATA2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 66.0 | 226.0 | 113.0 | 95.0 |
| 02 | 62.0 | 52.0 | 79.0 | 307.0 |
| 03 | 40.0 | 25.0 | 326.0 | 109.0 |
| 04 | 66.0 | 111.0 | 207.0 | 116.0 |
| 05 | 136.0 | 78.0 | 145.0 | 141.0 |
| 06 | 152.0 | 79.0 | 153.0 | 116.0 |
| 07 | 124.0 | 99.0 | 184.0 | 93.0 |
| 08 | 135.0 | 98.0 | 138.0 | 129.0 |
| 09 | 111.0 | 112.0 | 134.0 | 143.0 |
| 10 | 163.0 | 57.0 | 172.0 | 108.0 |
| 11 | 96.0 | 169.0 | 186.0 | 49.0 |
| 12 | 387.0 | 2.0 | 4.0 | 107.0 |
| 13 | 0.0 | 0.0 | 498.0 | 2.0 |
| 14 | 493.0 | 0.0 | 4.0 | 3.0 |
| 15 | 3.0 | 1.0 | 8.0 | 488.0 |
| 16 | 483.0 | 3.0 | 0.0 | 14.0 |
| 17 | 422.0 | 5.0 | 57.0 | 16.0 |
| 18 | 70.0 | 84.0 | 314.0 | 32.0 |
| 19 | 172.0 | 96.0 | 195.0 | 37.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.628 | 0.587 | -0.1 | -0.271 |
| 02 | -0.689 | -0.86 | -0.452 | 0.891 |
| 03 | -1.114 | -1.561 | 0.951 | -0.135 |
| 04 | -0.628 | -0.117 | 0.5 | -0.074 |
| 05 | 0.083 | -0.464 | 0.147 | 0.119 |
| 06 | 0.193 | -0.452 | 0.2 | -0.074 |
| 07 | -0.008 | -0.23 | 0.383 | -0.291 |
| 08 | 0.076 | -0.24 | 0.098 | 0.031 |
| 09 | -0.117 | -0.108 | 0.069 | 0.133 |
| 10 | 0.263 | -0.771 | 0.316 | -0.144 |
| 11 | -0.26 | 0.298 | 0.393 | -0.918 |
| 12 | 1.122 | -3.573 | -3.126 | -0.153 |
| 13 | -4.4 | -4.4 | 1.373 | -3.573 |
| 14 | 1.363 | -4.4 | -3.126 | -3.325 |
| 15 | -3.325 | -3.903 | -2.584 | 1.353 |
| 16 | 1.343 | -3.325 | -4.4 | -2.096 |
| 17 | 1.208 | -2.961 | -0.771 | -1.975 |
| 18 | -0.57 | -0.392 | 0.914 | -1.328 |
| 19 | 0.316 | -0.26 | 0.44 | -1.189 |