Model info
| Transcription factor | Gata4 | ||||||||
| Model | GATA4_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 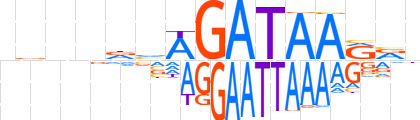 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndnndvWGATAARvn | ||||||||
| Best auROC (human) | 0.947 | ||||||||
| Best auROC (mouse) | 0.949 | ||||||||
| Peak sets in benchmark (human) | 15 | ||||||||
| Peak sets in benchmark (mouse) | 17 | ||||||||
| Aligned words | 428 | ||||||||
| TF family | GATA-type zinc fingers {2.2.1} | ||||||||
| TF subfamily | Two zinc-finger GATA factors {2.2.1.1} | ||||||||
| MGI | MGI:95664 | ||||||||
| EntrezGene | GeneID:14463 (SSTAR profile) | ||||||||
| UniProt ID | GATA4_MOUSE | ||||||||
| UniProt AC | Q08369 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Gata4 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 14.0 | 19.0 | 70.0 | 27.0 | 15.0 | 16.0 | 3.0 | 27.0 | 29.0 | 18.0 | 53.0 | 12.0 | 22.0 | 26.0 | 51.0 | 22.0 |
| 02 | 27.0 | 9.0 | 26.0 | 18.0 | 30.0 | 11.0 | 4.0 | 34.0 | 59.0 | 31.0 | 52.0 | 35.0 | 18.0 | 9.0 | 27.0 | 34.0 |
| 03 | 28.0 | 16.0 | 63.0 | 27.0 | 25.0 | 13.0 | 3.0 | 19.0 | 40.0 | 15.0 | 35.0 | 19.0 | 22.0 | 13.0 | 52.0 | 34.0 |
| 04 | 53.0 | 10.0 | 34.0 | 18.0 | 40.0 | 5.0 | 4.0 | 8.0 | 65.0 | 13.0 | 64.0 | 11.0 | 45.0 | 4.0 | 33.0 | 17.0 |
| 05 | 59.0 | 69.0 | 60.0 | 15.0 | 11.0 | 14.0 | 3.0 | 4.0 | 23.0 | 64.0 | 38.0 | 10.0 | 24.0 | 15.0 | 12.0 | 3.0 |
| 06 | 99.0 | 1.0 | 2.0 | 15.0 | 93.0 | 6.0 | 0.0 | 63.0 | 88.0 | 1.0 | 3.0 | 21.0 | 27.0 | 0.0 | 1.0 | 4.0 |
| 07 | 1.0 | 0.0 | 306.0 | 0.0 | 1.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 0.0 | 0.0 | 103.0 | 0.0 |
| 08 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 421.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 2.0 | 3.0 | 7.0 | 411.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 391.0 | 3.0 | 10.0 | 8.0 |
| 11 | 378.0 | 9.0 | 11.0 | 3.0 | 3.0 | 0.0 | 0.0 | 0.0 | 10.0 | 1.0 | 1.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 |
| 12 | 61.0 | 52.0 | 270.0 | 16.0 | 1.0 | 3.0 | 6.0 | 0.0 | 3.0 | 2.0 | 7.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 |
| 13 | 20.0 | 13.0 | 26.0 | 6.0 | 39.0 | 6.0 | 7.0 | 5.0 | 145.0 | 45.0 | 93.0 | 3.0 | 4.0 | 5.0 | 7.0 | 0.0 |
| 14 | 52.0 | 20.0 | 60.0 | 76.0 | 22.0 | 20.0 | 10.0 | 17.0 | 35.0 | 33.0 | 30.0 | 35.0 | 3.0 | 6.0 | 2.0 | 3.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.626 | -0.327 | 0.963 | 0.018 | -0.558 | -0.495 | -2.074 | 0.018 | 0.089 | -0.38 | 0.686 | -0.775 | -0.183 | -0.019 | 0.648 | -0.183 |
| 02 | 0.018 | -1.053 | -0.019 | -0.38 | 0.122 | -0.86 | -1.815 | 0.246 | 0.793 | 0.155 | 0.667 | 0.275 | -0.38 | -1.053 | 0.018 | 0.246 |
| 03 | 0.054 | -0.495 | 0.858 | 0.018 | -0.057 | -0.698 | -2.074 | -0.327 | 0.407 | -0.558 | 0.275 | -0.327 | -0.183 | -0.698 | 0.667 | 0.246 |
| 04 | 0.686 | -0.952 | 0.246 | -0.38 | 0.407 | -1.609 | -1.815 | -1.166 | 0.889 | -0.698 | 0.873 | -0.86 | 0.524 | -1.815 | 0.217 | -0.436 |
| 05 | 0.793 | 0.948 | 0.809 | -0.558 | -0.86 | -0.626 | -2.074 | -1.815 | -0.14 | 0.873 | 0.356 | -0.952 | -0.098 | -0.558 | -0.775 | -2.074 |
| 06 | 1.308 | -2.971 | -2.425 | -0.558 | 1.245 | -1.438 | -4.264 | 0.858 | 1.19 | -2.971 | -2.074 | -0.229 | 0.018 | -4.264 | -2.971 | -1.815 |
| 07 | -2.971 | -4.264 | 2.434 | -4.264 | -2.971 | -4.264 | -1.293 | -4.264 | -4.264 | -4.264 | -1.438 | -4.264 | -4.264 | -4.264 | 1.347 | -4.264 |
| 08 | -2.425 | -4.264 | -4.264 | -4.264 | -4.264 | -4.264 | -4.264 | -4.264 | 2.752 | -4.264 | -2.971 | -4.264 | -4.264 | -4.264 | -4.264 | -4.264 |
| 09 | -2.425 | -2.074 | -1.293 | 2.728 | -4.264 | -4.264 | -4.264 | -4.264 | -4.264 | -4.264 | -4.264 | -2.971 | -4.264 | -4.264 | -4.264 | -4.264 |
| 10 | -2.425 | -4.264 | -4.264 | -4.264 | -2.971 | -4.264 | -2.425 | -4.264 | -1.293 | -4.264 | -4.264 | -4.264 | 2.678 | -2.074 | -0.952 | -1.166 |
| 11 | 2.645 | -1.053 | -0.86 | -2.074 | -2.074 | -4.264 | -4.264 | -4.264 | -0.952 | -2.971 | -2.971 | -4.264 | -1.166 | -4.264 | -4.264 | -4.264 |
| 12 | 0.826 | 0.667 | 2.309 | -0.495 | -2.971 | -2.074 | -1.438 | -4.264 | -2.074 | -2.425 | -1.293 | -4.264 | -4.264 | -4.264 | -2.074 | -4.264 |
| 13 | -0.277 | -0.698 | -0.019 | -1.438 | 0.382 | -1.438 | -1.293 | -1.609 | 1.688 | 0.524 | 1.245 | -2.074 | -1.815 | -1.609 | -1.293 | -4.264 |
| 14 | 0.667 | -0.277 | 0.809 | 1.044 | -0.183 | -0.277 | -0.952 | -0.436 | 0.275 | 0.217 | 0.122 | 0.275 | -2.074 | -1.438 | -2.425 | -2.074 |