Model info
| Transcription factor | Hnf4a | ||||||||
| Model | HNF4A_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 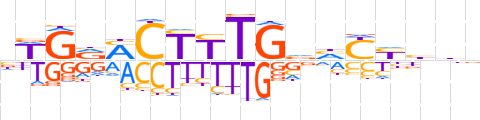 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | hhvRRdbCARAGKbCWn | ||||||||
| Best auROC (human) | 0.987 | ||||||||
| Best auROC (mouse) | 0.988 | ||||||||
| Peak sets in benchmark (human) | 49 | ||||||||
| Peak sets in benchmark (mouse) | 33 | ||||||||
| Aligned words | 386 | ||||||||
| TF family | RXR-related receptors (NR2) {2.1.3} | ||||||||
| TF subfamily | HNF-4 (NR2A) {2.1.3.2} | ||||||||
| MGI | MGI:109128 | ||||||||
| EntrezGene | GeneID:15378 (SSTAR profile) | ||||||||
| UniProt ID | HNF4A_MOUSE | ||||||||
| UniProt AC | P49698 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Hnf4a expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 27.0 | 25.0 | 20.0 | 8.0 | 89.0 | 35.0 | 7.0 | 29.0 | 20.0 | 14.0 | 16.0 | 11.0 | 19.0 | 23.0 | 23.0 | 20.0 |
| 02 | 67.0 | 26.0 | 48.0 | 14.0 | 51.0 | 22.0 | 9.0 | 15.0 | 26.0 | 14.0 | 22.0 | 4.0 | 13.0 | 10.0 | 39.0 | 6.0 |
| 03 | 81.0 | 7.0 | 61.0 | 8.0 | 52.0 | 2.0 | 4.0 | 14.0 | 54.0 | 9.0 | 44.0 | 11.0 | 14.0 | 2.0 | 20.0 | 3.0 |
| 04 | 19.0 | 4.0 | 171.0 | 7.0 | 6.0 | 1.0 | 4.0 | 9.0 | 13.0 | 13.0 | 87.0 | 16.0 | 1.0 | 1.0 | 29.0 | 5.0 |
| 05 | 13.0 | 4.0 | 15.0 | 7.0 | 4.0 | 0.0 | 1.0 | 14.0 | 41.0 | 14.0 | 91.0 | 145.0 | 3.0 | 1.0 | 14.0 | 19.0 |
| 06 | 4.0 | 17.0 | 28.0 | 12.0 | 3.0 | 3.0 | 1.0 | 12.0 | 19.0 | 24.0 | 45.0 | 33.0 | 14.0 | 116.0 | 21.0 | 34.0 |
| 07 | 2.0 | 36.0 | 0.0 | 2.0 | 7.0 | 141.0 | 0.0 | 12.0 | 0.0 | 81.0 | 6.0 | 8.0 | 2.0 | 79.0 | 4.0 | 6.0 |
| 08 | 7.0 | 1.0 | 3.0 | 0.0 | 320.0 | 8.0 | 5.0 | 4.0 | 9.0 | 1.0 | 0.0 | 0.0 | 21.0 | 0.0 | 7.0 | 0.0 |
| 09 | 256.0 | 10.0 | 77.0 | 14.0 | 6.0 | 0.0 | 4.0 | 0.0 | 11.0 | 0.0 | 4.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 |
| 10 | 220.0 | 7.0 | 42.0 | 8.0 | 10.0 | 0.0 | 0.0 | 0.0 | 65.0 | 4.0 | 10.0 | 6.0 | 12.0 | 0.0 | 2.0 | 0.0 |
| 11 | 16.0 | 6.0 | 277.0 | 8.0 | 4.0 | 0.0 | 3.0 | 4.0 | 0.0 | 2.0 | 50.0 | 2.0 | 0.0 | 0.0 | 13.0 | 1.0 |
| 12 | 4.0 | 1.0 | 13.0 | 2.0 | 6.0 | 1.0 | 0.0 | 1.0 | 23.0 | 11.0 | 56.0 | 253.0 | 1.0 | 4.0 | 2.0 | 8.0 |
| 13 | 4.0 | 11.0 | 10.0 | 9.0 | 5.0 | 5.0 | 1.0 | 6.0 | 3.0 | 14.0 | 18.0 | 36.0 | 23.0 | 191.0 | 14.0 | 36.0 |
| 14 | 3.0 | 29.0 | 3.0 | 0.0 | 6.0 | 191.0 | 3.0 | 21.0 | 5.0 | 32.0 | 4.0 | 2.0 | 6.0 | 69.0 | 4.0 | 8.0 |
| 15 | 5.0 | 5.0 | 8.0 | 2.0 | 249.0 | 23.0 | 13.0 | 36.0 | 11.0 | 3.0 | 0.0 | 0.0 | 11.0 | 4.0 | 12.0 | 4.0 |
| 16 | 62.0 | 79.0 | 93.0 | 42.0 | 15.0 | 8.0 | 4.0 | 8.0 | 9.0 | 16.0 | 2.0 | 6.0 | 8.0 | 9.0 | 11.0 | 14.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.111 | 0.035 | -0.184 | -1.074 | 1.294 | 0.367 | -1.201 | 0.181 | -0.184 | -0.533 | -0.403 | -0.767 | -0.235 | -0.047 | -0.047 | -0.184 |
| 02 | 1.012 | 0.074 | 0.68 | -0.533 | 0.741 | -0.091 | -0.961 | -0.466 | 0.074 | -0.533 | -0.091 | -1.723 | -0.605 | -0.859 | 0.475 | -1.347 |
| 03 | 1.2 | -1.201 | 0.918 | -1.074 | 0.76 | -2.335 | -1.723 | -0.533 | 0.797 | -0.961 | 0.594 | -0.767 | -0.533 | -2.335 | -0.184 | -1.983 |
| 04 | -0.235 | -1.723 | 1.945 | -1.201 | -1.347 | -2.882 | -1.723 | -0.961 | -0.605 | -0.605 | 1.272 | -0.403 | -2.882 | -2.882 | 0.181 | -1.517 |
| 05 | -0.605 | -1.723 | -0.466 | -1.201 | -1.723 | -4.187 | -2.882 | -0.533 | 0.524 | -0.533 | 1.316 | 1.781 | -1.983 | -2.882 | -0.533 | -0.235 |
| 06 | -1.723 | -0.344 | 0.147 | -0.683 | -1.983 | -1.983 | -2.882 | -0.683 | -0.235 | -0.005 | 0.616 | 0.309 | -0.533 | 1.558 | -0.136 | 0.339 |
| 07 | -2.335 | 0.395 | -4.187 | -2.335 | -1.201 | 1.753 | -4.187 | -0.683 | -4.187 | 1.2 | -1.347 | -1.074 | -2.335 | 1.176 | -1.723 | -1.347 |
| 08 | -1.201 | -2.882 | -1.983 | -4.187 | 2.571 | -1.074 | -1.517 | -1.723 | -0.961 | -2.882 | -4.187 | -4.187 | -0.136 | -4.187 | -1.201 | -4.187 |
| 09 | 2.348 | -0.859 | 1.15 | -0.533 | -1.347 | -4.187 | -1.723 | -4.187 | -0.767 | -4.187 | -1.723 | -4.187 | -1.723 | -4.187 | -4.187 | -4.187 |
| 10 | 2.197 | -1.201 | 0.548 | -1.074 | -0.859 | -4.187 | -4.187 | -4.187 | 0.982 | -1.723 | -0.859 | -1.347 | -0.683 | -4.187 | -2.335 | -4.187 |
| 11 | -0.403 | -1.347 | 2.427 | -1.074 | -1.723 | -4.187 | -1.983 | -1.723 | -4.187 | -2.335 | 0.721 | -2.335 | -4.187 | -4.187 | -0.605 | -2.882 |
| 12 | -1.723 | -2.882 | -0.605 | -2.335 | -1.347 | -2.882 | -4.187 | -2.882 | -0.047 | -0.767 | 0.833 | 2.336 | -2.882 | -1.723 | -2.335 | -1.074 |
| 13 | -1.723 | -0.767 | -0.859 | -0.961 | -1.517 | -1.517 | -2.882 | -1.347 | -1.983 | -0.533 | -0.288 | 0.395 | -0.047 | 2.056 | -0.533 | 0.395 |
| 14 | -1.983 | 0.181 | -1.983 | -4.187 | -1.347 | 2.056 | -1.983 | -0.136 | -1.517 | 0.279 | -1.723 | -2.335 | -1.347 | 1.041 | -1.723 | -1.074 |
| 15 | -1.517 | -1.517 | -1.074 | -2.335 | 2.32 | -0.047 | -0.605 | 0.395 | -0.767 | -1.983 | -4.187 | -4.187 | -0.767 | -1.723 | -0.683 | -1.723 |
| 16 | 0.935 | 1.176 | 1.338 | 0.548 | -0.466 | -1.074 | -1.723 | -1.074 | -0.961 | -0.403 | -2.335 | -1.347 | -1.074 | -0.961 | -0.767 | -0.533 |