Model info
| Transcription factor | HOXA9 (GeneCards) | ||||||||
| Model | HXA9_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 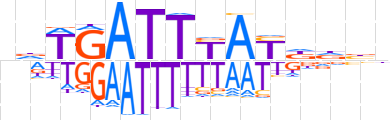 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndTGATTYAYddbh | ||||||||
| Best auROC (human) | 0.914 | ||||||||
| Best auROC (mouse) | 0.76 | ||||||||
| Peak sets in benchmark (human) | 5 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 502 | ||||||||
| TF family | HOX-related factors {3.1.1} | ||||||||
| TF subfamily | HOX9-13 {3.1.1.8} | ||||||||
| HGNC | HGNC:5109 | ||||||||
| EntrezGene | GeneID:3205 (SSTAR profile) | ||||||||
| UniProt ID | HXA9_HUMAN | ||||||||
| UniProt AC | P31269 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | HOXA9 expression | ||||||||
| ReMap ChIP-seq dataset list | HOXA9 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 49.0 | 14.0 | 41.0 | 18.0 | 65.0 | 8.0 | 2.0 | 14.0 | 77.0 | 12.0 | 26.0 | 20.0 | 38.0 | 11.0 | 50.0 | 38.0 |
| 02 | 54.0 | 0.0 | 7.0 | 168.0 | 9.0 | 0.0 | 0.0 | 36.0 | 15.0 | 2.0 | 5.0 | 97.0 | 7.0 | 0.0 | 8.0 | 75.0 |
| 03 | 9.0 | 0.0 | 71.0 | 5.0 | 2.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 13.0 | 2.0 | 43.0 | 2.0 | 316.0 | 15.0 |
| 04 | 55.0 | 0.0 | 0.0 | 4.0 | 2.0 | 0.0 | 0.0 | 0.0 | 399.0 | 1.0 | 0.0 | 0.0 | 22.0 | 0.0 | 0.0 | 0.0 |
| 05 | 9.0 | 2.0 | 7.0 | 460.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 |
| 06 | 3.0 | 0.0 | 0.0 | 6.0 | 0.0 | 0.0 | 0.0 | 2.0 | 1.0 | 0.0 | 0.0 | 6.0 | 21.0 | 0.0 | 9.0 | 435.0 |
| 07 | 3.0 | 0.0 | 2.0 | 20.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 3.0 | 1.0 | 21.0 | 52.0 | 41.0 | 335.0 |
| 08 | 28.0 | 1.0 | 0.0 | 0.0 | 35.0 | 16.0 | 1.0 | 0.0 | 46.0 | 0.0 | 0.0 | 0.0 | 341.0 | 7.0 | 8.0 | 0.0 |
| 09 | 7.0 | 68.0 | 25.0 | 350.0 | 18.0 | 3.0 | 2.0 | 1.0 | 0.0 | 2.0 | 1.0 | 6.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 1.0 | 0.0 | 3.0 | 21.0 | 30.0 | 0.0 | 11.0 | 32.0 | 7.0 | 1.0 | 4.0 | 16.0 | 59.0 | 15.0 | 203.0 | 80.0 |
| 11 | 15.0 | 4.0 | 65.0 | 13.0 | 5.0 | 0.0 | 2.0 | 9.0 | 77.0 | 14.0 | 86.0 | 44.0 | 28.0 | 7.0 | 72.0 | 42.0 |
| 12 | 5.0 | 63.0 | 27.0 | 30.0 | 8.0 | 8.0 | 1.0 | 8.0 | 29.0 | 116.0 | 31.0 | 49.0 | 17.0 | 50.0 | 21.0 | 20.0 |
| 13 | 6.0 | 21.0 | 18.0 | 14.0 | 50.0 | 78.0 | 13.0 | 96.0 | 11.0 | 19.0 | 19.0 | 31.0 | 17.0 | 27.0 | 27.0 | 36.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.48 | -0.754 | 0.303 | -0.509 | 0.76 | -1.294 | -2.55 | -0.754 | 0.929 | -0.904 | -0.147 | -0.405 | 0.228 | -0.988 | 0.5 | 0.228 |
| 02 | 0.576 | -4.371 | -1.421 | 1.706 | -1.181 | -4.371 | -4.371 | 0.174 | -0.687 | -2.55 | -1.736 | 1.159 | -1.421 | -4.371 | -1.294 | 0.902 |
| 03 | -1.181 | -4.371 | 0.848 | -1.736 | -2.55 | -4.371 | -4.371 | -4.371 | -1.736 | -4.371 | -0.826 | -2.55 | 0.35 | -2.55 | 2.337 | -0.687 |
| 04 | 0.594 | -4.371 | -4.371 | -1.942 | -2.55 | -4.371 | -4.371 | -4.371 | 2.57 | -3.094 | -4.371 | -4.371 | -0.312 | -4.371 | -4.371 | -4.371 |
| 05 | -1.181 | -2.55 | -1.421 | 2.712 | -4.371 | -4.371 | -4.371 | -3.094 | -4.371 | -4.371 | -4.371 | -4.371 | -4.371 | -4.371 | -4.371 | -1.942 |
| 06 | -2.2 | -4.371 | -4.371 | -1.566 | -4.371 | -4.371 | -4.371 | -2.55 | -3.094 | -4.371 | -4.371 | -1.566 | -0.357 | -4.371 | -1.181 | 2.656 |
| 07 | -2.2 | -4.371 | -2.55 | -0.405 | -4.371 | -4.371 | -4.371 | -4.371 | -1.736 | -4.371 | -2.2 | -3.094 | -0.357 | 0.539 | 0.303 | 2.395 |
| 08 | -0.074 | -3.094 | -4.371 | -4.371 | 0.146 | -0.624 | -3.094 | -4.371 | 0.417 | -4.371 | -4.371 | -4.371 | 2.413 | -1.421 | -1.294 | -4.371 |
| 09 | -1.421 | 0.805 | -0.186 | 2.439 | -0.509 | -2.2 | -2.55 | -3.094 | -4.371 | -2.55 | -3.094 | -1.566 | -4.371 | -4.371 | -4.371 | -4.371 |
| 10 | -3.094 | -4.371 | -2.2 | -0.357 | -0.006 | -4.371 | -0.988 | 0.058 | -1.421 | -3.094 | -1.942 | -0.624 | 0.664 | -0.687 | 1.895 | 0.967 |
| 11 | -0.687 | -1.942 | 0.76 | -0.826 | -1.736 | -4.371 | -2.55 | -1.181 | 0.929 | -0.754 | 1.039 | 0.373 | -0.074 | -1.421 | 0.862 | 0.327 |
| 12 | -1.736 | 0.729 | -0.11 | -0.006 | -1.294 | -1.294 | -3.094 | -1.294 | -0.04 | 1.337 | 0.026 | 0.48 | -0.564 | 0.5 | -0.357 | -0.405 |
| 13 | -1.566 | -0.357 | -0.509 | -0.754 | 0.5 | 0.942 | -0.826 | 1.148 | -0.988 | -0.456 | -0.456 | 0.026 | -0.564 | -0.11 | -0.11 | 0.174 |