Model info
| Transcription factor | HOXC12 (GeneCards) | ||||||||
| Model | HXC12_HUMAN.H11MO.0.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 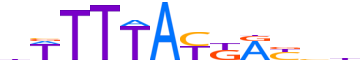 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | HT-SELEX | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 12 | ||||||||
| Quality | D | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dWTTTAYKRYbb | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 2466 | ||||||||
| TF family | HOX-related factors {3.1.1} | ||||||||
| TF subfamily | HOX9-13 {3.1.1.8} | ||||||||
| HGNC | HGNC:5124 | ||||||||
| EntrezGene | GeneID:3228 (SSTAR profile) | ||||||||
| UniProt ID | HXC12_HUMAN | ||||||||
| UniProt AC | P31275 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | HOXC12 expression | ||||||||
| ReMap ChIP-seq dataset list | HOXC12 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 742.75 | 285.75 | 546.75 | 889.75 |
| 02 | 279.25 | 143.25 | 251.25 | 1791.25 |
| 03 | 39.5 | 20.5 | 25.5 | 2379.5 |
| 04 | 30.75 | 6.75 | 6.75 | 2420.75 |
| 05 | 353.0 | 4.0 | 22.0 | 2086.0 |
| 06 | 2453.0 | 1.0 | 2.0 | 9.0 |
| 07 | 5.0 | 1207.0 | 15.0 | 1238.0 |
| 08 | 90.0 | 108.0 | 1343.0 | 924.0 |
| 09 | 1819.0 | 75.0 | 457.0 | 114.0 |
| 10 | 135.0 | 1467.0 | 328.0 | 535.0 |
| 11 | 384.0 | 880.0 | 402.0 | 799.0 |
| 12 | 409.5 | 464.5 | 465.5 | 1125.5 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.186 | -0.765 | -0.119 | 0.366 |
| 02 | -0.788 | -1.449 | -0.893 | 1.065 |
| 03 | -2.702 | -3.315 | -3.114 | 1.349 |
| 04 | -2.939 | -4.263 | -4.263 | 1.366 |
| 05 | -0.555 | -4.643 | -3.251 | 1.217 |
| 06 | 1.379 | -5.344 | -5.052 | -4.033 |
| 07 | -4.488 | 0.671 | -3.596 | 0.696 |
| 08 | -1.906 | -1.727 | 0.777 | 0.404 |
| 09 | 1.08 | -2.084 | -0.298 | -1.674 |
| 10 | -1.507 | 0.865 | -0.628 | -0.141 |
| 11 | -0.471 | 0.355 | -0.426 | 0.259 |
| 12 | -0.407 | -0.282 | -0.28 | 0.601 |