Model info
| Transcription factor | Irf8 | ||||||||
| Model | IRF8_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 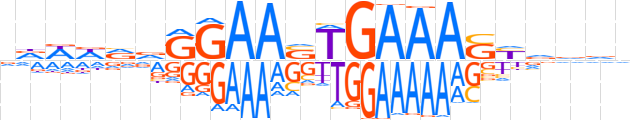 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ddWWvdGGAASTGAAASYvvvd | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.997 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 59 | ||||||||
| Aligned words | 513 | ||||||||
| TF family | Interferon-regulatory factors {3.5.3} | ||||||||
| TF subfamily | IRF-8 (ICSBP1) {3.5.3.0.8} | ||||||||
| MGI | MGI:96395 | ||||||||
| EntrezGene | GeneID:15900 (SSTAR profile) | ||||||||
| UniProt ID | IRF8_MOUSE | ||||||||
| UniProt AC | P23611 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Irf8 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 77.0 | 10.0 | 38.0 | 14.0 | 37.0 | 6.0 | 2.0 | 10.0 | 79.0 | 9.0 | 18.0 | 18.0 | 22.0 | 7.0 | 11.0 | 22.0 |
| 02 | 159.0 | 6.0 | 26.0 | 24.0 | 19.0 | 4.0 | 3.0 | 6.0 | 49.0 | 4.0 | 12.0 | 4.0 | 14.0 | 4.0 | 13.0 | 33.0 |
| 03 | 149.0 | 14.0 | 37.0 | 41.0 | 12.0 | 0.0 | 2.0 | 4.0 | 33.0 | 3.0 | 9.0 | 9.0 | 21.0 | 2.0 | 9.0 | 35.0 |
| 04 | 57.0 | 26.0 | 121.0 | 11.0 | 15.0 | 1.0 | 1.0 | 2.0 | 27.0 | 8.0 | 18.0 | 4.0 | 27.0 | 6.0 | 50.0 | 6.0 |
| 05 | 50.0 | 11.0 | 56.0 | 9.0 | 30.0 | 0.0 | 6.0 | 5.0 | 84.0 | 28.0 | 51.0 | 27.0 | 8.0 | 1.0 | 13.0 | 1.0 |
| 06 | 9.0 | 2.0 | 161.0 | 0.0 | 32.0 | 0.0 | 8.0 | 0.0 | 40.0 | 2.0 | 84.0 | 0.0 | 4.0 | 6.0 | 29.0 | 3.0 |
| 07 | 9.0 | 0.0 | 76.0 | 0.0 | 7.0 | 0.0 | 3.0 | 0.0 | 42.0 | 0.0 | 240.0 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 |
| 08 | 56.0 | 0.0 | 2.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 321.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 372.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 10 | 54.0 | 98.0 | 214.0 | 9.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 |
| 11 | 1.0 | 5.0 | 0.0 | 48.0 | 21.0 | 7.0 | 0.0 | 70.0 | 23.0 | 5.0 | 0.0 | 191.0 | 0.0 | 1.0 | 0.0 | 8.0 |
| 12 | 0.0 | 0.0 | 45.0 | 0.0 | 0.0 | 0.0 | 18.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 315.0 | 1.0 |
| 13 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 370.0 | 1.0 | 7.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 14 | 357.0 | 0.0 | 14.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 363.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.0 | 1.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 16 | 10.0 | 143.0 | 221.0 | 2.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 |
| 17 | 1.0 | 0.0 | 1.0 | 8.0 | 21.0 | 35.0 | 0.0 | 89.0 | 34.0 | 47.0 | 16.0 | 126.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 18 | 20.0 | 2.0 | 32.0 | 2.0 | 50.0 | 11.0 | 12.0 | 10.0 | 8.0 | 1.0 | 9.0 | 0.0 | 56.0 | 37.0 | 100.0 | 30.0 |
| 19 | 43.0 | 20.0 | 49.0 | 22.0 | 21.0 | 9.0 | 10.0 | 11.0 | 72.0 | 22.0 | 42.0 | 17.0 | 8.0 | 9.0 | 22.0 | 3.0 |
| 20 | 78.0 | 18.0 | 39.0 | 9.0 | 23.0 | 15.0 | 2.0 | 20.0 | 45.0 | 24.0 | 42.0 | 12.0 | 11.0 | 5.0 | 20.0 | 17.0 |
| 21 | 94.0 | 18.0 | 27.0 | 18.0 | 20.0 | 14.0 | 3.0 | 25.0 | 42.0 | 18.0 | 27.0 | 16.0 | 16.0 | 8.0 | 14.0 | 20.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.166 | -0.844 | 0.464 | -0.518 | 0.438 | -1.331 | -2.32 | -0.844 | 1.191 | -0.945 | -0.272 | -0.272 | -0.075 | -1.186 | -0.752 | -0.075 |
| 02 | 1.888 | -1.331 | 0.089 | 0.01 | -0.219 | -1.708 | -1.968 | -1.331 | 0.716 | -1.708 | -0.668 | -1.708 | -0.518 | -1.708 | -0.59 | 0.325 |
| 03 | 1.823 | -0.518 | 0.438 | 0.539 | -0.668 | -4.174 | -2.32 | -1.708 | 0.325 | -1.968 | -0.945 | -0.945 | -0.121 | -2.32 | -0.945 | 0.383 |
| 04 | 0.866 | 0.089 | 1.616 | -0.752 | -0.451 | -2.867 | -2.867 | -2.32 | 0.126 | -1.058 | -0.272 | -1.708 | 0.126 | -1.331 | 0.736 | -1.331 |
| 05 | 0.736 | -0.752 | 0.849 | -0.945 | 0.23 | -4.174 | -1.331 | -1.502 | 1.252 | 0.162 | 0.756 | 0.126 | -1.058 | -2.867 | -0.59 | -2.867 |
| 06 | -0.945 | -2.32 | 1.901 | -4.174 | 0.294 | -4.174 | -1.058 | -4.174 | 0.515 | -2.32 | 1.252 | -4.174 | -1.708 | -1.331 | 0.197 | -1.968 |
| 07 | -0.945 | -4.174 | 1.153 | -4.174 | -1.186 | -4.174 | -1.968 | -4.174 | 0.563 | -4.174 | 2.299 | -4.174 | -2.867 | -4.174 | -2.32 | -4.174 |
| 08 | 0.849 | -4.174 | -2.32 | -2.867 | -4.174 | -4.174 | -4.174 | -4.174 | 2.59 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 |
| 09 | 2.737 | -4.174 | -4.174 | -1.502 | -4.174 | -4.174 | -4.174 | -4.174 | -2.32 | -4.174 | -4.174 | -4.174 | -2.867 | -4.174 | -4.174 | -4.174 |
| 10 | 0.813 | 1.406 | 2.185 | -0.945 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -1.502 | -4.174 |
| 11 | -2.867 | -1.502 | -4.174 | 0.696 | -0.121 | -1.186 | -4.174 | 1.071 | -0.032 | -1.502 | -4.174 | 2.071 | -4.174 | -2.867 | -4.174 | -1.058 |
| 12 | -4.174 | -4.174 | 0.632 | -4.174 | -4.174 | -4.174 | -0.272 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -2.867 | -4.174 | 2.571 | -2.867 |
| 13 | -2.867 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | 2.731 | -2.867 | -1.186 | -4.174 | -2.867 | -4.174 | -4.174 | -4.174 |
| 14 | 2.696 | -4.174 | -0.518 | -2.867 | -2.867 | -4.174 | -4.174 | -4.174 | -1.186 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 |
| 15 | 2.712 | -4.174 | -4.174 | -2.32 | -4.174 | -4.174 | -4.174 | -4.174 | -0.668 | -2.867 | -4.174 | -2.867 | -2.867 | -4.174 | -4.174 | -4.174 |
| 16 | -0.844 | 1.782 | 2.217 | -2.32 | -4.174 | -2.867 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -4.174 | -2.867 | -2.32 | -4.174 |
| 17 | -2.867 | -4.174 | -2.867 | -1.058 | -0.121 | 0.383 | -4.174 | 1.31 | 0.354 | 0.675 | -0.388 | 1.656 | -4.174 | -2.867 | -2.867 | -4.174 |
| 18 | -0.169 | -2.32 | 0.294 | -2.32 | 0.736 | -0.752 | -0.668 | -0.844 | -1.058 | -2.867 | -0.945 | -4.174 | 0.849 | 0.438 | 1.426 | 0.23 |
| 19 | 0.587 | -0.169 | 0.716 | -0.075 | -0.121 | -0.945 | -0.844 | -0.752 | 1.099 | -0.075 | 0.563 | -0.328 | -1.058 | -0.945 | -0.075 | -1.968 |
| 20 | 1.178 | -0.272 | 0.49 | -0.945 | -0.032 | -0.451 | -2.32 | -0.169 | 0.632 | 0.01 | 0.563 | -0.668 | -0.752 | -1.502 | -0.169 | -0.328 |
| 21 | 1.364 | -0.272 | 0.126 | -0.272 | -0.169 | -0.518 | -1.968 | 0.051 | 0.563 | -0.272 | 0.126 | -0.388 | -0.388 | -1.058 | -0.518 | -0.169 |