Model info
| Transcription factor | TCF4 (GeneCards) | ||||||||
| Model | ITF2_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 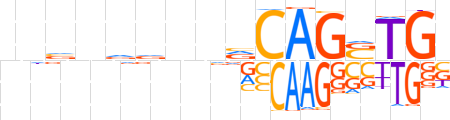 | ||||||||
| LOGO (reverse complement) | 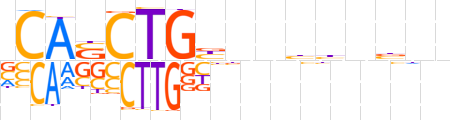 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vnvnvvnnvCAGSTGb | ||||||||
| Best auROC (human) | 0.968 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 471 | ||||||||
| TF family | E2A-related factors {1.2.1} | ||||||||
| TF subfamily | SEF2 (E2-2, TCF-4, ITF-2) {1.2.1.0.2} | ||||||||
| HGNC | HGNC:11634 | ||||||||
| EntrezGene | GeneID:6925 (SSTAR profile) | ||||||||
| UniProt ID | ITF2_HUMAN | ||||||||
| UniProt AC | P15884 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | TCF4 expression | ||||||||
| ReMap ChIP-seq dataset list | TCF4 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 52.0 | 9.0 | 35.0 | 7.0 | 67.0 | 35.0 | 10.0 | 74.0 | 33.0 | 15.0 | 35.0 | 31.0 | 8.0 | 8.0 | 34.0 | 17.0 |
| 02 | 31.0 | 38.0 | 83.0 | 8.0 | 26.0 | 27.0 | 4.0 | 10.0 | 18.0 | 32.0 | 52.0 | 12.0 | 11.0 | 24.0 | 80.0 | 14.0 |
| 03 | 12.0 | 22.0 | 42.0 | 10.0 | 41.0 | 44.0 | 12.0 | 24.0 | 47.0 | 63.0 | 56.0 | 53.0 | 6.0 | 20.0 | 8.0 | 10.0 |
| 04 | 26.0 | 7.0 | 63.0 | 10.0 | 71.0 | 27.0 | 26.0 | 25.0 | 39.0 | 21.0 | 49.0 | 9.0 | 19.0 | 11.0 | 58.0 | 9.0 |
| 05 | 29.0 | 14.0 | 96.0 | 16.0 | 25.0 | 14.0 | 11.0 | 16.0 | 51.0 | 30.0 | 85.0 | 30.0 | 3.0 | 13.0 | 30.0 | 7.0 |
| 06 | 32.0 | 13.0 | 45.0 | 18.0 | 32.0 | 14.0 | 9.0 | 16.0 | 52.0 | 63.0 | 75.0 | 32.0 | 5.0 | 14.0 | 31.0 | 19.0 |
| 07 | 31.0 | 30.0 | 45.0 | 15.0 | 30.0 | 40.0 | 6.0 | 28.0 | 46.0 | 37.0 | 58.0 | 19.0 | 17.0 | 20.0 | 33.0 | 15.0 |
| 08 | 29.0 | 20.0 | 73.0 | 2.0 | 71.0 | 36.0 | 16.0 | 4.0 | 26.0 | 51.0 | 63.0 | 2.0 | 17.0 | 5.0 | 54.0 | 1.0 |
| 09 | 5.0 | 133.0 | 4.0 | 1.0 | 2.0 | 109.0 | 1.0 | 0.0 | 3.0 | 199.0 | 4.0 | 0.0 | 1.0 | 8.0 | 0.0 | 0.0 |
| 10 | 8.0 | 0.0 | 3.0 | 0.0 | 435.0 | 1.0 | 11.0 | 2.0 | 7.0 | 0.0 | 2.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 11 | 2.0 | 18.0 | 425.0 | 6.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 14.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 12 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 17.0 | 3.0 | 0.0 | 53.0 | 232.0 | 153.0 | 4.0 | 0.0 | 5.0 | 1.0 | 0.0 |
| 13 | 4.0 | 1.0 | 2.0 | 46.0 | 13.0 | 4.0 | 5.0 | 234.0 | 8.0 | 0.0 | 4.0 | 145.0 | 1.0 | 0.0 | 0.0 | 3.0 |
| 14 | 1.0 | 0.0 | 24.0 | 1.0 | 1.0 | 0.0 | 1.0 | 3.0 | 2.0 | 1.0 | 8.0 | 0.0 | 3.0 | 2.0 | 415.0 | 8.0 |
| 15 | 3.0 | 1.0 | 3.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 37.0 | 161.0 | 146.0 | 104.0 | 2.0 | 4.0 | 2.0 | 4.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.565 | -1.154 | 0.173 | -1.394 | 0.817 | 0.173 | -1.053 | 0.916 | 0.115 | -0.66 | 0.173 | 0.053 | -1.267 | -1.267 | 0.144 | -0.538 |
| 02 | 0.053 | 0.255 | 1.03 | -1.267 | -0.12 | -0.083 | -1.915 | -1.053 | -0.482 | 0.085 | 0.565 | -0.877 | -0.961 | -0.199 | 0.994 | -0.727 |
| 03 | -0.877 | -0.285 | 0.354 | -1.053 | 0.33 | 0.4 | -0.877 | -0.199 | 0.465 | 0.756 | 0.639 | 0.584 | -1.539 | -0.378 | -1.267 | -1.053 |
| 04 | -0.12 | -1.394 | 0.756 | -1.053 | 0.875 | -0.083 | -0.12 | -0.159 | 0.28 | -0.33 | 0.506 | -1.154 | -0.429 | -0.961 | 0.674 | -1.154 |
| 05 | -0.013 | -0.727 | 1.175 | -0.597 | -0.159 | -0.727 | -0.961 | -0.597 | 0.546 | 0.021 | 1.054 | 0.021 | -2.174 | -0.799 | 0.021 | -1.394 |
| 06 | 0.085 | -0.799 | 0.422 | -0.482 | 0.085 | -0.727 | -1.154 | -0.597 | 0.565 | 0.756 | 0.929 | 0.085 | -1.71 | -0.727 | 0.053 | -0.429 |
| 07 | 0.053 | 0.021 | 0.422 | -0.66 | 0.021 | 0.305 | -1.539 | -0.047 | 0.444 | 0.228 | 0.674 | -0.429 | -0.538 | -0.378 | 0.115 | -0.66 |
| 08 | -0.013 | -0.378 | 0.903 | -2.524 | 0.875 | 0.201 | -0.597 | -1.915 | -0.12 | 0.546 | 0.756 | -2.524 | -0.538 | -1.71 | 0.603 | -3.068 |
| 09 | -1.71 | 1.5 | -1.915 | -3.068 | -2.524 | 1.302 | -3.068 | -4.349 | -2.174 | 1.902 | -1.915 | -4.349 | -3.068 | -1.267 | -4.349 | -4.349 |
| 10 | -1.267 | -4.349 | -2.174 | -4.349 | 2.683 | -3.068 | -0.961 | -2.524 | -1.394 | -4.349 | -2.524 | -4.349 | -3.068 | -4.349 | -4.349 | -4.349 |
| 11 | -2.524 | -0.482 | 2.66 | -1.539 | -4.349 | -4.349 | -3.068 | -4.349 | -4.349 | -2.524 | -0.727 | -4.349 | -4.349 | -4.349 | -2.524 | -4.349 |
| 12 | -4.349 | -2.524 | -4.349 | -4.349 | -4.349 | -0.538 | -2.174 | -4.349 | 0.584 | 2.055 | 1.64 | -1.915 | -4.349 | -1.71 | -3.068 | -4.349 |
| 13 | -1.915 | -3.068 | -2.524 | 0.444 | -0.799 | -1.915 | -1.71 | 2.064 | -1.267 | -4.349 | -1.915 | 1.586 | -3.068 | -4.349 | -4.349 | -2.174 |
| 14 | -3.068 | -4.349 | -0.199 | -3.068 | -3.068 | -4.349 | -3.068 | -2.174 | -2.524 | -3.068 | -1.267 | -4.349 | -2.174 | -2.524 | 2.636 | -1.267 |
| 15 | -2.174 | -3.068 | -2.174 | -4.349 | -4.349 | -3.068 | -3.068 | -3.068 | 0.228 | 1.691 | 1.593 | 1.255 | -2.524 | -1.915 | -2.524 | -1.915 |