Model info
| Transcription factor | ZBTB33 (GeneCards) | ||||||||
| Model | KAISO_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 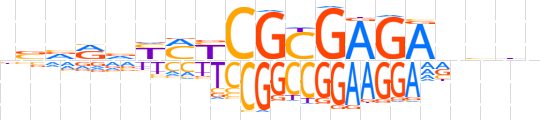 | ||||||||
| LOGO (reverse complement) | 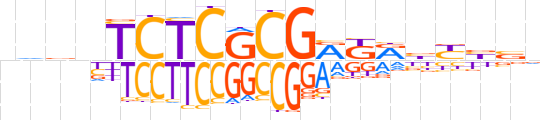 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbvRvYMYCGCGAGAvbhv | ||||||||
| Best auROC (human) | 0.904 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 37 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 408 | ||||||||
| TF family | Other factors with up to three adjacent zinc fingers {2.3.2} | ||||||||
| TF subfamily | Factors with 2-3 adjacent zinc fingersand a BTB/POZ domain {2.3.2.1} | ||||||||
| HGNC | HGNC:16682 | ||||||||
| EntrezGene | GeneID:10009 (SSTAR profile) | ||||||||
| UniProt ID | KAISO_HUMAN | ||||||||
| UniProt AC | Q86T24 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZBTB33 expression | ||||||||
| ReMap ChIP-seq dataset list | ZBTB33 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.0 | 21.0 | 25.0 | 4.0 | 17.0 | 66.0 | 38.0 | 13.0 | 15.0 | 53.0 | 42.0 | 15.0 | 3.0 | 60.0 | 27.0 | 5.0 |
| 02 | 18.0 | 8.0 | 7.0 | 3.0 | 115.0 | 36.0 | 36.0 | 13.0 | 69.0 | 18.0 | 29.0 | 16.0 | 12.0 | 9.0 | 16.0 | 0.0 |
| 03 | 85.0 | 13.0 | 114.0 | 2.0 | 29.0 | 6.0 | 32.0 | 4.0 | 29.0 | 11.0 | 46.0 | 2.0 | 10.0 | 8.0 | 12.0 | 2.0 |
| 04 | 55.0 | 22.0 | 61.0 | 15.0 | 6.0 | 15.0 | 10.0 | 7.0 | 123.0 | 33.0 | 34.0 | 14.0 | 2.0 | 3.0 | 4.0 | 1.0 |
| 05 | 18.0 | 28.0 | 18.0 | 122.0 | 1.0 | 17.0 | 4.0 | 51.0 | 8.0 | 41.0 | 4.0 | 56.0 | 2.0 | 6.0 | 1.0 | 28.0 |
| 06 | 12.0 | 8.0 | 8.0 | 1.0 | 45.0 | 40.0 | 5.0 | 2.0 | 8.0 | 16.0 | 2.0 | 1.0 | 62.0 | 183.0 | 12.0 | 0.0 |
| 07 | 5.0 | 27.0 | 12.0 | 83.0 | 11.0 | 27.0 | 28.0 | 181.0 | 3.0 | 3.0 | 4.0 | 17.0 | 0.0 | 1.0 | 2.0 | 1.0 |
| 08 | 0.0 | 17.0 | 2.0 | 0.0 | 2.0 | 54.0 | 1.0 | 1.0 | 2.0 | 43.0 | 1.0 | 0.0 | 1.0 | 280.0 | 1.0 | 0.0 |
| 09 | 0.0 | 1.0 | 4.0 | 0.0 | 19.0 | 0.0 | 373.0 | 2.0 | 0.0 | 0.0 | 5.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 10 | 3.0 | 8.0 | 3.0 | 6.0 | 0.0 | 1.0 | 0.0 | 0.0 | 12.0 | 321.0 | 4.0 | 45.0 | 1.0 | 1.0 | 0.0 | 0.0 |
| 11 | 6.0 | 0.0 | 10.0 | 0.0 | 4.0 | 0.0 | 327.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 2.0 | 0.0 | 47.0 | 2.0 |
| 12 | 5.0 | 1.0 | 1.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 356.0 | 0.0 | 30.0 | 5.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 13 | 9.0 | 7.0 | 337.0 | 8.0 | 0.0 | 0.0 | 1.0 | 0.0 | 4.0 | 1.0 | 26.0 | 0.0 | 2.0 | 0.0 | 10.0 | 0.0 |
| 14 | 10.0 | 3.0 | 2.0 | 0.0 | 7.0 | 1.0 | 0.0 | 0.0 | 334.0 | 11.0 | 25.0 | 4.0 | 6.0 | 1.0 | 1.0 | 0.0 |
| 15 | 128.0 | 82.0 | 104.0 | 43.0 | 3.0 | 3.0 | 7.0 | 3.0 | 10.0 | 4.0 | 9.0 | 5.0 | 0.0 | 2.0 | 0.0 | 2.0 |
| 16 | 14.0 | 75.0 | 38.0 | 14.0 | 6.0 | 23.0 | 13.0 | 49.0 | 18.0 | 40.0 | 33.0 | 29.0 | 10.0 | 10.0 | 5.0 | 28.0 |
| 17 | 12.0 | 6.0 | 17.0 | 13.0 | 27.0 | 23.0 | 25.0 | 73.0 | 38.0 | 20.0 | 17.0 | 14.0 | 13.0 | 29.0 | 10.0 | 68.0 |
| 18 | 14.0 | 21.0 | 44.0 | 11.0 | 13.0 | 25.0 | 27.0 | 13.0 | 23.0 | 18.0 | 19.0 | 9.0 | 31.0 | 73.0 | 40.0 | 24.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -2.927 | -0.184 | -0.012 | -1.77 | -0.391 | 0.949 | 0.401 | -0.653 | -0.513 | 0.731 | 0.501 | -0.513 | -2.03 | 0.855 | 0.064 | -1.564 |
| 02 | -0.335 | -1.121 | -1.248 | -2.03 | 1.502 | 0.348 | 0.348 | -0.653 | 0.994 | -0.335 | 0.134 | -0.45 | -0.73 | -1.008 | -0.45 | -4.226 |
| 03 | 1.201 | -0.653 | 1.493 | -2.381 | 0.134 | -1.394 | 0.231 | -1.77 | 0.134 | -0.815 | 0.591 | -2.381 | -0.907 | -1.121 | -0.73 | -2.381 |
| 04 | 0.768 | -0.138 | 0.871 | -0.513 | -1.394 | -0.513 | -0.907 | -1.248 | 1.569 | 0.262 | 0.291 | -0.581 | -2.381 | -2.03 | -1.77 | -2.927 |
| 05 | -0.335 | 0.1 | -0.335 | 1.561 | -2.927 | -0.391 | -1.77 | 0.693 | -1.121 | 0.477 | -1.77 | 0.786 | -2.381 | -1.394 | -2.927 | 0.1 |
| 06 | -0.73 | -1.121 | -1.121 | -2.927 | 0.569 | 0.452 | -1.564 | -2.381 | -1.121 | -0.45 | -2.381 | -2.927 | 0.887 | 1.966 | -0.73 | -4.226 |
| 07 | -1.564 | 0.064 | -0.73 | 1.177 | -0.815 | 0.064 | 0.1 | 1.955 | -2.03 | -2.03 | -1.77 | -0.391 | -4.226 | -2.927 | -2.381 | -2.927 |
| 08 | -4.226 | -0.391 | -2.381 | -4.226 | -2.381 | 0.75 | -2.927 | -2.927 | -2.381 | 0.524 | -2.927 | -4.226 | -2.927 | 2.39 | -2.927 | -4.226 |
| 09 | -4.226 | -2.927 | -1.77 | -4.226 | -0.282 | -4.226 | 2.677 | -2.381 | -4.226 | -4.226 | -1.564 | -4.226 | -2.927 | -4.226 | -4.226 | -4.226 |
| 10 | -2.03 | -1.121 | -2.03 | -1.394 | -4.226 | -2.927 | -4.226 | -4.226 | -0.73 | 2.527 | -1.77 | 0.569 | -2.927 | -2.927 | -4.226 | -4.226 |
| 11 | -1.394 | -4.226 | -0.907 | -4.226 | -1.77 | -4.226 | 2.545 | -4.226 | -4.226 | -4.226 | -1.248 | -4.226 | -2.381 | -4.226 | 0.612 | -2.381 |
| 12 | -1.564 | -2.927 | -2.927 | -1.564 | -4.226 | -4.226 | -4.226 | -4.226 | 2.63 | -4.226 | 0.168 | -1.564 | -4.226 | -4.226 | -4.226 | -2.381 |
| 13 | -1.008 | -1.248 | 2.575 | -1.121 | -4.226 | -4.226 | -2.927 | -4.226 | -1.77 | -2.927 | 0.026 | -4.226 | -2.381 | -4.226 | -0.907 | -4.226 |
| 14 | -0.907 | -2.03 | -2.381 | -4.226 | -1.248 | -2.927 | -4.226 | -4.226 | 2.566 | -0.815 | -0.012 | -1.77 | -1.394 | -2.927 | -2.927 | -4.226 |
| 15 | 1.609 | 1.165 | 1.402 | 0.524 | -2.03 | -2.03 | -1.248 | -2.03 | -0.907 | -1.77 | -1.008 | -1.564 | -4.226 | -2.381 | -4.226 | -2.381 |
| 16 | -0.581 | 1.076 | 0.401 | -0.581 | -1.394 | -0.094 | -0.653 | 0.653 | -0.335 | 0.452 | 0.262 | 0.134 | -0.907 | -0.907 | -1.564 | 0.1 |
| 17 | -0.73 | -1.394 | -0.391 | -0.653 | 0.064 | -0.094 | -0.012 | 1.05 | 0.401 | -0.232 | -0.391 | -0.581 | -0.653 | 0.134 | -0.907 | 0.979 |
| 18 | -0.581 | -0.184 | 0.547 | -0.815 | -0.653 | -0.012 | 0.064 | -0.653 | -0.094 | -0.335 | -0.282 | -1.008 | 0.2 | 1.05 | 0.452 | -0.052 |