Model info
| Transcription factor | Klf15 | ||||||||
| Model | KLF15_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 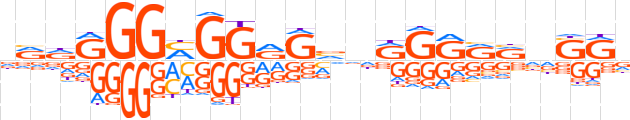 | ||||||||
| LOGO (reverse complement) | 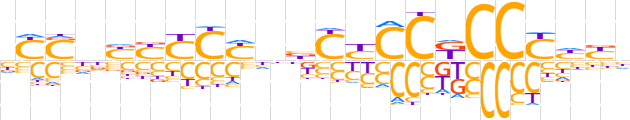 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dRdRGGMGGRGvdRGRRRdSRv | ||||||||
| Best auROC (human) | 0.967 | ||||||||
| Best auROC (mouse) | 0.998 | ||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 530 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | Krüppel-like factors {2.3.1.2} | ||||||||
| MGI | MGI:1929988 | ||||||||
| EntrezGene | GeneID:66277 (SSTAR profile) | ||||||||
| UniProt ID | KLF15_MOUSE | ||||||||
| UniProt AC | Q9EPW2 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Klf15 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 27.0 | 8.0 | 93.0 | 5.0 | 19.0 | 6.0 | 34.0 | 7.0 | 74.0 | 39.0 | 103.0 | 6.0 | 11.0 | 9.0 | 50.0 | 6.0 |
| 02 | 39.0 | 3.0 | 79.0 | 10.0 | 16.0 | 2.0 | 32.0 | 12.0 | 77.0 | 21.0 | 136.0 | 46.0 | 6.0 | 2.0 | 13.0 | 3.0 |
| 03 | 13.0 | 0.0 | 102.0 | 23.0 | 2.0 | 0.0 | 25.0 | 1.0 | 97.0 | 3.0 | 159.0 | 1.0 | 1.0 | 0.0 | 70.0 | 0.0 |
| 04 | 1.0 | 0.0 | 112.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 1.0 | 0.0 | 355.0 | 0.0 | 0.0 | 0.0 | 25.0 | 0.0 |
| 05 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 3.0 | 485.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 06 | 4.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 228.0 | 207.0 | 0.0 | 54.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 07 | 37.0 | 0.0 | 196.0 | 0.0 | 2.0 | 0.0 | 204.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 55.0 | 0.0 |
| 08 | 2.0 | 0.0 | 14.0 | 24.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 399.0 | 53.0 | 0.0 | 0.0 | 1.0 | 1.0 |
| 09 | 2.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 195.0 | 9.0 | 186.0 | 24.0 | 6.0 | 11.0 | 56.0 | 5.0 |
| 10 | 11.0 | 5.0 | 184.0 | 3.0 | 1.0 | 1.0 | 15.0 | 3.0 | 48.0 | 6.0 | 173.0 | 17.0 | 2.0 | 0.0 | 26.0 | 2.0 |
| 11 | 35.0 | 13.0 | 13.0 | 1.0 | 3.0 | 2.0 | 6.0 | 1.0 | 141.0 | 144.0 | 94.0 | 19.0 | 9.0 | 11.0 | 5.0 | 0.0 |
| 12 | 75.0 | 21.0 | 74.0 | 18.0 | 46.0 | 22.0 | 47.0 | 55.0 | 58.0 | 23.0 | 6.0 | 31.0 | 2.0 | 1.0 | 14.0 | 4.0 |
| 13 | 30.0 | 3.0 | 116.0 | 32.0 | 11.0 | 5.0 | 34.0 | 17.0 | 36.0 | 2.0 | 95.0 | 8.0 | 7.0 | 5.0 | 86.0 | 10.0 |
| 14 | 5.0 | 1.0 | 76.0 | 2.0 | 2.0 | 3.0 | 7.0 | 3.0 | 46.0 | 2.0 | 272.0 | 11.0 | 6.0 | 1.0 | 56.0 | 4.0 |
| 15 | 15.0 | 0.0 | 43.0 | 1.0 | 2.0 | 0.0 | 5.0 | 0.0 | 113.0 | 16.0 | 277.0 | 5.0 | 1.0 | 2.0 | 16.0 | 1.0 |
| 16 | 14.0 | 4.0 | 112.0 | 1.0 | 3.0 | 4.0 | 9.0 | 2.0 | 62.0 | 60.0 | 204.0 | 15.0 | 1.0 | 0.0 | 4.0 | 2.0 |
| 17 | 36.0 | 2.0 | 40.0 | 2.0 | 2.0 | 3.0 | 61.0 | 2.0 | 40.0 | 61.0 | 203.0 | 25.0 | 1.0 | 2.0 | 14.0 | 3.0 |
| 18 | 18.0 | 1.0 | 31.0 | 29.0 | 8.0 | 8.0 | 48.0 | 4.0 | 127.0 | 64.0 | 72.0 | 55.0 | 2.0 | 2.0 | 26.0 | 2.0 |
| 19 | 8.0 | 12.0 | 130.0 | 5.0 | 5.0 | 0.0 | 67.0 | 3.0 | 16.0 | 14.0 | 120.0 | 27.0 | 2.0 | 22.0 | 60.0 | 6.0 |
| 20 | 5.0 | 1.0 | 21.0 | 4.0 | 22.0 | 1.0 | 17.0 | 8.0 | 34.0 | 2.0 | 298.0 | 43.0 | 3.0 | 1.0 | 33.0 | 4.0 |
| 21 | 4.0 | 12.0 | 27.0 | 21.0 | 1.0 | 1.0 | 1.0 | 2.0 | 106.0 | 75.0 | 145.0 | 43.0 | 3.0 | 16.0 | 31.0 | 9.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.138 | -1.322 | 1.088 | -1.764 | -0.484 | -1.594 | 0.089 | -1.449 | 0.861 | 0.225 | 1.19 | -1.594 | -1.016 | -1.209 | 0.471 | -1.594 |
| 02 | 0.225 | -2.228 | 0.926 | -1.108 | -0.652 | -2.578 | 0.029 | -0.932 | 0.9 | -0.386 | 1.467 | 0.389 | -1.594 | -2.578 | -0.854 | -2.228 |
| 03 | -0.854 | -4.395 | 1.18 | -0.296 | -2.578 | -4.395 | -0.214 | -3.121 | 1.13 | -2.228 | 1.623 | -3.121 | -3.121 | -4.395 | 0.806 | -4.395 |
| 04 | -3.121 | -4.395 | 1.274 | -4.395 | -4.395 | -4.395 | -3.121 | -2.578 | -3.121 | -4.395 | 2.425 | -4.395 | -4.395 | -4.395 | -0.214 | -4.395 |
| 05 | -4.395 | -4.395 | -2.578 | -4.395 | -4.395 | -4.395 | -4.395 | -4.395 | -1.97 | -2.228 | 2.737 | -3.121 | -4.395 | -4.395 | -2.578 | -4.395 |
| 06 | -1.97 | -4.395 | -4.395 | -4.395 | -3.121 | -4.395 | -4.395 | -2.578 | 1.983 | 1.886 | -4.395 | 0.548 | -4.395 | -3.121 | -4.395 | -4.395 |
| 07 | 0.173 | -4.395 | 1.832 | -4.395 | -2.578 | -4.395 | 1.872 | -2.578 | -4.395 | -4.395 | -4.395 | -4.395 | -3.121 | -4.395 | 0.566 | -4.395 |
| 08 | -2.578 | -4.395 | -0.782 | -0.254 | -4.395 | -4.395 | -4.395 | -4.395 | -3.121 | -2.578 | 2.542 | 0.529 | -4.395 | -4.395 | -3.121 | -3.121 |
| 09 | -2.578 | -4.395 | -4.395 | -3.121 | -4.395 | -4.395 | -2.578 | -4.395 | 1.827 | -1.209 | 1.779 | -0.254 | -1.594 | -1.016 | 0.584 | -1.764 |
| 10 | -1.016 | -1.764 | 1.769 | -2.228 | -3.121 | -3.121 | -0.715 | -2.228 | 0.431 | -1.594 | 1.707 | -0.593 | -2.578 | -4.395 | -0.176 | -2.578 |
| 11 | 0.118 | -0.854 | -0.854 | -3.121 | -2.228 | -2.578 | -1.594 | -3.121 | 1.503 | 1.524 | 1.099 | -0.484 | -1.209 | -1.016 | -1.764 | -4.395 |
| 12 | 0.874 | -0.386 | 0.861 | -0.537 | 0.389 | -0.34 | 0.41 | 0.566 | 0.619 | -0.296 | -1.594 | -0.002 | -2.578 | -3.121 | -0.782 | -1.97 |
| 13 | -0.034 | -2.228 | 1.309 | 0.029 | -1.016 | -1.764 | 0.089 | -0.593 | 0.146 | -2.578 | 1.11 | -1.322 | -1.449 | -1.764 | 1.01 | -1.108 |
| 14 | -1.764 | -3.121 | 0.887 | -2.578 | -2.578 | -2.228 | -1.449 | -2.228 | 0.389 | -2.578 | 2.159 | -1.016 | -1.594 | -3.121 | 0.584 | -1.97 |
| 15 | -0.715 | -4.395 | 0.322 | -3.121 | -2.578 | -4.395 | -1.764 | -4.395 | 1.282 | -0.652 | 2.177 | -1.764 | -3.121 | -2.578 | -0.652 | -3.121 |
| 16 | -0.782 | -1.97 | 1.274 | -3.121 | -2.228 | -1.97 | -1.209 | -2.578 | 0.685 | 0.652 | 1.872 | -0.715 | -3.121 | -4.395 | -1.97 | -2.578 |
| 17 | 0.146 | -2.578 | 0.25 | -2.578 | -2.578 | -2.228 | 0.669 | -2.578 | 0.25 | 0.669 | 1.867 | -0.214 | -3.121 | -2.578 | -0.782 | -2.228 |
| 18 | -0.537 | -3.121 | -0.002 | -0.068 | -1.322 | -1.322 | 0.431 | -1.97 | 1.399 | 0.717 | 0.834 | 0.566 | -2.578 | -2.578 | -0.176 | -2.578 |
| 19 | -1.322 | -0.932 | 1.422 | -1.764 | -1.764 | -4.395 | 0.762 | -2.228 | -0.652 | -0.782 | 1.342 | -0.138 | -2.578 | -0.34 | 0.652 | -1.594 |
| 20 | -1.764 | -3.121 | -0.386 | -1.97 | -0.34 | -3.121 | -0.593 | -1.322 | 0.089 | -2.578 | 2.25 | 0.322 | -2.228 | -3.121 | 0.06 | -1.97 |
| 21 | -1.97 | -0.932 | -0.138 | -0.386 | -3.121 | -3.121 | -3.121 | -2.578 | 1.219 | 0.874 | 1.531 | 0.322 | -2.228 | -0.652 | -0.002 | -1.209 |