Model info
| Transcription factor | KLF4 (GeneCards) | ||||||||
| Model | KLF4_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 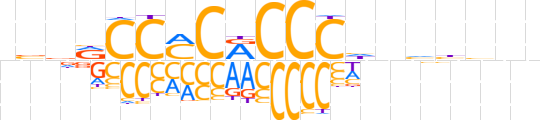 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvddbndGGGYGKGGYhvn | ||||||||
| Best auROC (human) | 0.904 | ||||||||
| Best auROC (mouse) | 0.987 | ||||||||
| Peak sets in benchmark (human) | 11 | ||||||||
| Peak sets in benchmark (mouse) | 34 | ||||||||
| Aligned words | 455 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | Krüppel-like factors {2.3.1.2} | ||||||||
| HGNC | HGNC:6348 | ||||||||
| EntrezGene | GeneID:9314 (SSTAR profile) | ||||||||
| UniProt ID | KLF4_HUMAN | ||||||||
| UniProt AC | O43474 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | KLF4 expression | ||||||||
| ReMap ChIP-seq dataset list | KLF4 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 41.0 | 14.0 | 43.0 | 18.0 | 25.0 | 12.0 | 20.0 | 22.0 | 63.0 | 22.0 | 73.0 | 22.0 | 10.0 | 28.0 | 29.0 | 12.0 |
| 02 | 25.0 | 10.0 | 83.0 | 21.0 | 27.0 | 20.0 | 11.0 | 18.0 | 32.0 | 38.0 | 67.0 | 28.0 | 9.0 | 14.0 | 33.0 | 18.0 |
| 03 | 26.0 | 7.0 | 45.0 | 15.0 | 18.0 | 16.0 | 34.0 | 14.0 | 70.0 | 23.0 | 83.0 | 18.0 | 13.0 | 15.0 | 41.0 | 16.0 |
| 04 | 20.0 | 9.0 | 47.0 | 51.0 | 14.0 | 20.0 | 15.0 | 12.0 | 20.0 | 45.0 | 102.0 | 36.0 | 6.0 | 13.0 | 33.0 | 11.0 |
| 05 | 19.0 | 6.0 | 23.0 | 12.0 | 27.0 | 32.0 | 7.0 | 21.0 | 60.0 | 77.0 | 44.0 | 16.0 | 40.0 | 18.0 | 20.0 | 32.0 |
| 06 | 78.0 | 3.0 | 42.0 | 23.0 | 40.0 | 13.0 | 22.0 | 58.0 | 32.0 | 11.0 | 9.0 | 42.0 | 31.0 | 3.0 | 21.0 | 26.0 |
| 07 | 20.0 | 0.0 | 152.0 | 9.0 | 3.0 | 0.0 | 27.0 | 0.0 | 5.0 | 0.0 | 88.0 | 1.0 | 4.0 | 0.0 | 144.0 | 1.0 |
| 08 | 0.0 | 0.0 | 32.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 410.0 | 0.0 | 0.0 | 0.0 | 11.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 450.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 2.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 52.0 | 96.0 | 0.0 | 302.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 11 | 6.0 | 0.0 | 45.0 | 1.0 | 0.0 | 1.0 | 95.0 | 2.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 301.0 | 0.0 |
| 12 | 0.0 | 0.0 | 2.0 | 5.0 | 0.0 | 0.0 | 0.0 | 2.0 | 1.0 | 12.0 | 260.0 | 169.0 | 0.0 | 0.0 | 0.0 | 3.0 |
| 13 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 12.0 | 0.0 | 35.0 | 1.0 | 223.0 | 3.0 | 3.0 | 0.0 | 174.0 | 2.0 |
| 14 | 0.0 | 0.0 | 36.0 | 2.0 | 0.0 | 0.0 | 1.0 | 0.0 | 13.0 | 5.0 | 371.0 | 21.0 | 0.0 | 0.0 | 4.0 | 1.0 |
| 15 | 0.0 | 8.0 | 5.0 | 0.0 | 0.0 | 4.0 | 0.0 | 1.0 | 44.0 | 254.0 | 27.0 | 87.0 | 0.0 | 15.0 | 5.0 | 4.0 |
| 16 | 23.0 | 14.0 | 5.0 | 2.0 | 62.0 | 125.0 | 21.0 | 73.0 | 2.0 | 10.0 | 16.0 | 9.0 | 6.0 | 35.0 | 26.0 | 25.0 |
| 17 | 7.0 | 11.0 | 56.0 | 19.0 | 43.0 | 60.0 | 60.0 | 21.0 | 15.0 | 15.0 | 34.0 | 4.0 | 7.0 | 31.0 | 58.0 | 13.0 |
| 18 | 6.0 | 21.0 | 34.0 | 11.0 | 44.0 | 26.0 | 20.0 | 27.0 | 21.0 | 63.0 | 66.0 | 58.0 | 5.0 | 8.0 | 31.0 | 13.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.364 | -0.693 | 0.411 | -0.448 | -0.125 | -0.843 | -0.344 | -0.251 | 0.79 | -0.251 | 0.937 | -0.251 | -1.019 | -0.013 | 0.022 | -0.843 |
| 02 | -0.125 | -1.019 | 1.065 | -0.296 | -0.049 | -0.344 | -0.927 | -0.448 | 0.119 | 0.289 | 0.851 | -0.013 | -1.12 | -0.693 | 0.149 | -0.448 |
| 03 | -0.086 | -1.36 | 0.456 | -0.626 | -0.448 | -0.563 | 0.179 | -0.693 | 0.895 | -0.207 | 1.065 | -0.448 | -0.765 | -0.626 | 0.364 | -0.563 |
| 04 | -0.344 | -1.12 | 0.499 | 0.58 | -0.693 | -0.344 | -0.626 | -0.843 | -0.344 | 0.456 | 1.27 | 0.235 | -1.505 | -0.765 | 0.149 | -0.927 |
| 05 | -0.395 | -1.505 | -0.207 | -0.843 | -0.049 | 0.119 | -1.36 | -0.296 | 0.742 | 0.99 | 0.434 | -0.563 | 0.339 | -0.448 | -0.344 | 0.119 |
| 06 | 1.003 | -2.14 | 0.388 | -0.207 | 0.339 | -0.765 | -0.251 | 0.708 | 0.119 | -0.927 | -1.12 | 0.388 | 0.087 | -2.14 | -0.296 | -0.086 |
| 07 | -0.344 | -4.32 | 1.667 | -1.12 | -2.14 | -4.32 | -0.049 | -4.32 | -1.676 | -4.32 | 1.123 | -3.035 | -1.881 | -4.32 | 1.614 | -3.035 |
| 08 | -4.32 | -4.32 | 0.119 | -4.32 | -4.32 | -4.32 | -4.32 | -4.32 | -4.32 | -3.035 | 2.658 | -4.32 | -4.32 | -4.32 | -0.927 | -4.32 |
| 09 | -4.32 | -4.32 | -4.32 | -4.32 | -3.035 | -4.32 | -4.32 | -4.32 | -2.491 | -4.32 | 2.751 | -3.035 | -4.32 | -4.32 | -4.32 | -4.32 |
| 10 | -4.32 | -2.491 | -3.035 | -4.32 | -4.32 | -4.32 | -4.32 | -4.32 | 0.6 | 1.209 | -4.32 | 2.353 | -4.32 | -4.32 | -4.32 | -3.035 |
| 11 | -1.505 | -4.32 | 0.456 | -3.035 | -4.32 | -3.035 | 1.199 | -2.491 | -4.32 | -4.32 | -3.035 | -4.32 | -3.035 | -3.035 | 2.349 | -4.32 |
| 12 | -4.32 | -4.32 | -2.491 | -1.676 | -4.32 | -4.32 | -4.32 | -2.491 | -3.035 | -0.843 | 2.203 | 1.773 | -4.32 | -4.32 | -4.32 | -2.14 |
| 13 | -4.32 | -4.32 | -3.035 | -4.32 | -4.32 | -4.32 | -0.843 | -4.32 | 0.207 | -3.035 | 2.05 | -2.14 | -2.14 | -4.32 | 1.802 | -2.491 |
| 14 | -4.32 | -4.32 | 0.235 | -2.491 | -4.32 | -4.32 | -3.035 | -4.32 | -0.765 | -1.676 | 2.558 | -0.296 | -4.32 | -4.32 | -1.881 | -3.035 |
| 15 | -4.32 | -1.233 | -1.676 | -4.32 | -4.32 | -1.881 | -4.32 | -3.035 | 0.434 | 2.18 | -0.049 | 1.111 | -4.32 | -0.626 | -1.676 | -1.881 |
| 16 | -0.207 | -0.693 | -1.676 | -2.491 | 0.774 | 1.472 | -0.296 | 0.937 | -2.491 | -1.019 | -0.563 | -1.12 | -1.505 | 0.207 | -0.086 | -0.125 |
| 17 | -1.36 | -0.927 | 0.673 | -0.395 | 0.411 | 0.742 | 0.742 | -0.296 | -0.626 | -0.626 | 0.179 | -1.881 | -1.36 | 0.087 | 0.708 | -0.765 |
| 18 | -1.505 | -0.296 | 0.179 | -0.927 | 0.434 | -0.086 | -0.344 | -0.049 | -0.296 | 0.79 | 0.837 | 0.708 | -1.676 | -1.233 | 0.087 | -0.765 |