Model info
| Transcription factor | Lhx3 | ||||||||
| Model | LHX3_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 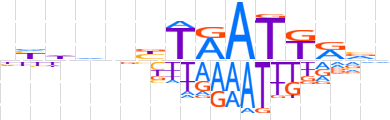 | ||||||||
| LOGO (reverse complement) | 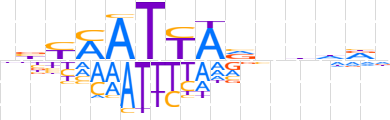 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | hhdhnbTAATKRvd | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.951 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 387 | ||||||||
| TF family | HD-LIM factors {3.1.5} | ||||||||
| TF subfamily | Lhx-3-like factors {3.1.5.4} | ||||||||
| MGI | MGI:102673 | ||||||||
| EntrezGene | GeneID:16871 (SSTAR profile) | ||||||||
| UniProt ID | LHX3_MOUSE | ||||||||
| UniProt AC | P50481 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Lhx3 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 6.0 | 16.0 | 11.0 | 49.0 | 6.0 | 11.0 | 2.0 | 62.0 | 10.0 | 16.0 | 7.0 | 14.0 | 25.0 | 32.0 | 17.0 | 88.0 |
| 02 | 16.0 | 3.0 | 16.0 | 12.0 | 22.0 | 11.0 | 2.0 | 40.0 | 13.0 | 4.0 | 8.0 | 12.0 | 41.0 | 29.0 | 34.0 | 109.0 |
| 03 | 41.0 | 17.0 | 18.0 | 16.0 | 17.0 | 8.0 | 4.0 | 18.0 | 25.0 | 20.0 | 8.0 | 7.0 | 67.0 | 36.0 | 27.0 | 43.0 |
| 04 | 36.0 | 24.0 | 53.0 | 37.0 | 30.0 | 16.0 | 6.0 | 29.0 | 20.0 | 11.0 | 20.0 | 6.0 | 12.0 | 17.0 | 46.0 | 9.0 |
| 05 | 0.0 | 45.0 | 36.0 | 17.0 | 5.0 | 38.0 | 1.0 | 24.0 | 8.0 | 72.0 | 15.0 | 30.0 | 1.0 | 17.0 | 21.0 | 42.0 |
| 06 | 0.0 | 0.0 | 0.0 | 14.0 | 45.0 | 0.0 | 0.0 | 127.0 | 11.0 | 0.0 | 0.0 | 62.0 | 15.0 | 0.0 | 0.0 | 98.0 |
| 07 | 42.0 | 0.0 | 29.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 199.0 | 0.0 | 102.0 | 0.0 |
| 08 | 240.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 129.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 1.0 | 0.0 | 35.0 | 333.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 10 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 31.0 | 4.0 | 0.0 | 132.0 | 200.0 |
| 11 | 1.0 | 0.0 | 2.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 74.0 | 13.0 | 45.0 | 4.0 | 113.0 | 6.0 | 107.0 | 6.0 |
| 12 | 76.0 | 33.0 | 71.0 | 8.0 | 15.0 | 2.0 | 2.0 | 0.0 | 52.0 | 60.0 | 36.0 | 6.0 | 5.0 | 2.0 | 4.0 | 0.0 |
| 13 | 66.0 | 10.0 | 27.0 | 45.0 | 25.0 | 20.0 | 3.0 | 49.0 | 39.0 | 22.0 | 24.0 | 28.0 | 2.0 | 4.0 | 5.0 | 3.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.311 | -0.367 | -0.731 | 0.737 | -1.311 | -0.731 | -2.299 | 0.971 | -0.823 | -0.367 | -1.165 | -0.497 | 0.071 | 0.315 | -0.307 | 1.319 |
| 02 | -0.367 | -1.947 | -0.367 | -0.647 | -0.054 | -0.731 | -2.299 | 0.536 | -0.569 | -1.687 | -1.037 | -0.647 | 0.56 | 0.218 | 0.375 | 1.533 |
| 03 | 0.56 | -0.307 | -0.251 | -0.367 | -0.307 | -1.037 | -1.687 | -0.251 | 0.071 | -0.148 | -1.037 | -1.165 | 1.048 | 0.432 | 0.147 | 0.608 |
| 04 | 0.432 | 0.031 | 0.815 | 0.459 | 0.251 | -0.367 | -1.311 | 0.218 | -0.148 | -0.731 | -0.148 | -1.311 | -0.647 | -0.307 | 0.675 | -0.925 |
| 05 | -4.157 | 0.653 | 0.432 | -0.307 | -1.481 | 0.485 | -2.847 | 0.031 | -1.037 | 1.12 | -0.43 | 0.251 | -2.847 | -0.307 | -0.1 | 0.584 |
| 06 | -4.157 | -4.157 | -4.157 | -0.497 | 0.653 | -4.157 | -4.157 | 1.685 | -0.731 | -4.157 | -4.157 | 0.971 | -0.43 | -4.157 | -4.157 | 1.427 |
| 07 | 0.584 | -4.157 | 0.218 | -4.157 | -4.157 | -4.157 | -4.157 | -4.157 | -4.157 | -4.157 | -4.157 | -4.157 | 2.133 | -4.157 | 1.467 | -4.157 |
| 08 | 2.32 | -4.157 | -4.157 | -2.847 | -4.157 | -4.157 | -4.157 | -4.157 | 1.701 | -2.847 | -2.847 | -4.157 | -4.157 | -4.157 | -4.157 | -4.157 |
| 09 | -2.847 | -4.157 | 0.404 | 2.647 | -4.157 | -4.157 | -4.157 | -2.847 | -4.157 | -4.157 | -4.157 | -2.847 | -4.157 | -4.157 | -4.157 | -2.847 |
| 10 | -4.157 | -4.157 | -4.157 | -2.847 | -4.157 | -4.157 | -4.157 | -4.157 | -4.157 | -4.157 | -1.687 | 0.284 | -1.687 | -4.157 | 1.724 | 2.138 |
| 11 | -2.847 | -4.157 | -2.299 | -2.847 | -4.157 | -4.157 | -4.157 | -4.157 | 1.147 | -0.569 | 0.653 | -1.687 | 1.569 | -1.311 | 1.514 | -1.311 |
| 12 | 1.173 | 0.346 | 1.106 | -1.037 | -0.43 | -2.299 | -2.299 | -4.157 | 0.796 | 0.938 | 0.432 | -1.311 | -1.481 | -2.299 | -1.687 | -4.157 |
| 13 | 1.033 | -0.823 | 0.147 | 0.653 | 0.071 | -0.148 | -1.947 | 0.737 | 0.511 | -0.054 | 0.031 | 0.183 | -2.299 | -1.687 | -1.481 | -1.947 |