Model info
| Transcription factor | LHX9 (GeneCards) | ||||||||
| Model | LHX9_HUMAN.H11MO.0.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 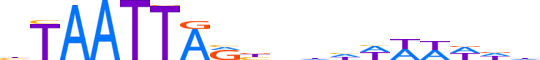 | ||||||||
| LOGO (reverse complement) | 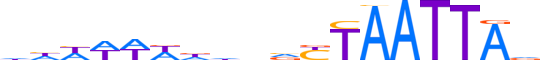 | ||||||||
| Data source | HT-SELEX | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 18 | ||||||||
| Quality | D | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bTAATTARbnhhWWWWdd | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 4409 | ||||||||
| TF family | HD-LIM factors {3.1.5} | ||||||||
| TF subfamily | Lhx-2-like factors {3.1.5.3} | ||||||||
| HGNC | HGNC:14222 | ||||||||
| EntrezGene | GeneID:56956 (SSTAR profile) | ||||||||
| UniProt ID | LHX9_HUMAN | ||||||||
| UniProt AC | Q9NQ69 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | LHX9 expression | ||||||||
| ReMap ChIP-seq dataset list | LHX9 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 567.0 | 1936.0 | 663.0 | 1235.0 |
| 02 | 64.75 | 407.75 | 59.75 | 3868.75 |
| 03 | 4326.0 | 69.0 | 2.0 | 4.0 |
| 04 | 4396.25 | 1.25 | 2.25 | 1.25 |
| 05 | 0.25 | 2.25 | 0.25 | 4398.25 |
| 06 | 2.25 | 7.25 | 95.25 | 4296.25 |
| 07 | 3527.25 | 1.25 | 870.25 | 2.25 |
| 08 | 951.25 | 614.25 | 2686.25 | 149.25 |
| 09 | 293.5 | 1896.5 | 700.5 | 1510.5 |
| 10 | 1053.75 | 1114.75 | 826.75 | 1405.75 |
| 11 | 1486.75 | 1009.75 | 484.75 | 1419.75 |
| 12 | 1588.5 | 679.5 | 293.5 | 1839.5 |
| 13 | 1679.5 | 341.5 | 132.5 | 2247.5 |
| 14 | 2240.5 | 177.5 | 46.5 | 1936.5 |
| 15 | 2269.5 | 118.5 | 149.5 | 1863.5 |
| 16 | 1743.5 | 184.5 | 349.5 | 2123.5 |
| 17 | 1463.25 | 310.25 | 663.25 | 1964.25 |
| 18 | 1497.5 | 501.5 | 923.5 | 1478.5 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.661 | 0.564 | -0.505 | 0.115 |
| 02 | -2.803 | -0.989 | -2.881 | 1.256 |
| 03 | 1.368 | -2.741 | -5.595 | -5.197 |
| 04 | 1.384 | -5.797 | -5.536 | -5.797 |
| 05 | -6.152 | -5.536 | -6.152 | 1.384 |
| 06 | -5.536 | -4.77 | -2.427 | 1.361 |
| 07 | 1.164 | -5.797 | -0.234 | -5.536 |
| 08 | -0.145 | -0.581 | 0.891 | -1.986 |
| 09 | -1.316 | 0.544 | -0.45 | 0.316 |
| 10 | -0.043 | 0.013 | -0.285 | 0.245 |
| 11 | 0.301 | -0.086 | -0.817 | 0.255 |
| 12 | 0.367 | -0.481 | -1.316 | 0.513 |
| 13 | 0.422 | -1.166 | -2.103 | 0.713 |
| 14 | 0.71 | -1.814 | -3.122 | 0.565 |
| 15 | 0.723 | -2.213 | -1.984 | 0.526 |
| 16 | 0.46 | -1.776 | -1.143 | 0.657 |
| 17 | 0.285 | -1.261 | -0.505 | 0.579 |
| 18 | 0.308 | -0.783 | -0.175 | 0.295 |