Model info
| Transcription factor | Lyl1 | ||||||||
| Model | LYL1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 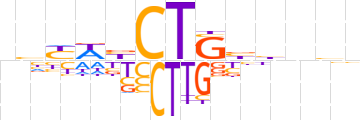 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nhYWbCTSbbnhn | ||||||||
| Best auROC (human) | 0.884 | ||||||||
| Best auROC (mouse) | 0.854 | ||||||||
| Peak sets in benchmark (human) | 23 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 476 | ||||||||
| TF family | Tal-related factors {1.2.3} | ||||||||
| TF subfamily | Tal / HEN-like factors {1.2.3.1} | ||||||||
| MGI | MGI:96891 | ||||||||
| EntrezGene | GeneID:17095 (SSTAR profile) | ||||||||
| UniProt ID | LYL1_MOUSE | ||||||||
| UniProt AC | P27792 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Lyl1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 39.0 | 61.0 | 20.0 | 22.0 | 28.0 | 49.0 | 2.0 | 50.0 | 11.0 | 64.0 | 23.0 | 30.0 | 16.0 | 33.0 | 19.0 | 9.0 |
| 02 | 8.0 | 66.0 | 12.0 | 8.0 | 21.0 | 111.0 | 5.0 | 70.0 | 5.0 | 41.0 | 2.0 | 16.0 | 4.0 | 28.0 | 3.0 | 76.0 |
| 03 | 10.0 | 4.0 | 8.0 | 16.0 | 180.0 | 9.0 | 15.0 | 42.0 | 8.0 | 0.0 | 1.0 | 13.0 | 66.0 | 1.0 | 16.0 | 87.0 |
| 04 | 12.0 | 12.0 | 77.0 | 163.0 | 2.0 | 2.0 | 1.0 | 9.0 | 2.0 | 18.0 | 8.0 | 12.0 | 3.0 | 86.0 | 24.0 | 45.0 |
| 05 | 0.0 | 19.0 | 0.0 | 0.0 | 0.0 | 118.0 | 0.0 | 0.0 | 0.0 | 107.0 | 1.0 | 2.0 | 0.0 | 220.0 | 7.0 | 2.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 2.0 | 460.0 | 0.0 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 | 4.0 |
| 07 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 5.0 | 57.0 | 364.0 | 46.0 |
| 08 | 2.0 | 0.0 | 0.0 | 3.0 | 7.0 | 23.0 | 0.0 | 27.0 | 19.0 | 133.0 | 78.0 | 138.0 | 1.0 | 19.0 | 4.0 | 22.0 |
| 09 | 3.0 | 5.0 | 5.0 | 16.0 | 40.0 | 29.0 | 7.0 | 99.0 | 13.0 | 25.0 | 21.0 | 23.0 | 17.0 | 41.0 | 46.0 | 86.0 |
| 10 | 24.0 | 15.0 | 19.0 | 15.0 | 16.0 | 35.0 | 4.0 | 45.0 | 5.0 | 26.0 | 23.0 | 25.0 | 44.0 | 43.0 | 42.0 | 95.0 |
| 11 | 34.0 | 26.0 | 8.0 | 21.0 | 34.0 | 48.0 | 7.0 | 30.0 | 16.0 | 28.0 | 13.0 | 31.0 | 15.0 | 89.0 | 36.0 | 40.0 |
| 12 | 23.0 | 19.0 | 43.0 | 14.0 | 36.0 | 97.0 | 7.0 | 51.0 | 9.0 | 18.0 | 23.0 | 14.0 | 8.0 | 33.0 | 44.0 | 37.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.268 | 0.711 | -0.391 | -0.297 | -0.06 | 0.494 | -2.536 | 0.514 | -0.973 | 0.759 | -0.254 | 0.008 | -0.609 | 0.102 | -0.441 | -1.167 |
| 02 | -1.279 | 0.79 | -0.889 | -1.279 | -0.343 | 1.307 | -1.722 | 0.848 | -1.722 | 0.317 | -2.536 | -0.609 | -1.927 | -0.06 | -2.186 | 0.93 |
| 03 | -1.065 | -1.927 | -1.279 | -0.609 | 1.789 | -1.167 | -0.672 | 0.341 | -1.279 | -4.359 | -3.08 | -0.812 | 0.79 | -3.08 | -0.609 | 1.065 |
| 04 | -0.889 | -0.889 | 0.943 | 1.69 | -2.536 | -2.536 | -3.08 | -1.167 | -2.536 | -0.494 | -1.279 | -0.889 | -2.186 | 1.053 | -0.212 | 0.409 |
| 05 | -4.359 | -0.441 | -4.359 | -4.359 | -4.359 | 1.368 | -4.359 | -4.359 | -4.359 | 1.271 | -3.08 | -2.536 | -4.359 | 1.99 | -1.406 | -2.536 |
| 06 | -4.359 | -4.359 | -4.359 | -4.359 | -3.08 | -3.08 | -2.536 | 2.726 | -4.359 | -4.359 | -4.359 | -1.279 | -4.359 | -4.359 | -4.359 | -1.927 |
| 07 | -4.359 | -4.359 | -3.08 | -4.359 | -4.359 | -4.359 | -3.08 | -4.359 | -4.359 | -4.359 | -2.536 | -4.359 | -1.722 | 0.644 | 2.493 | 0.431 |
| 08 | -2.536 | -4.359 | -4.359 | -2.186 | -1.406 | -0.254 | -4.359 | -0.096 | -0.441 | 1.488 | 0.956 | 1.524 | -3.08 | -0.441 | -1.927 | -0.297 |
| 09 | -2.186 | -1.722 | -1.722 | -0.609 | 0.293 | -0.025 | -1.406 | 1.193 | -0.812 | -0.172 | -0.343 | -0.254 | -0.55 | 0.317 | 0.431 | 1.053 |
| 10 | -0.212 | -0.672 | -0.441 | -0.672 | -0.609 | 0.161 | -1.927 | 0.409 | -1.722 | -0.133 | -0.254 | -0.172 | 0.387 | 0.364 | 0.341 | 1.152 |
| 11 | 0.132 | -0.133 | -1.279 | -0.343 | 0.132 | 0.473 | -1.406 | 0.008 | -0.609 | -0.06 | -0.812 | 0.041 | -0.672 | 1.087 | 0.188 | 0.293 |
| 12 | -0.254 | -0.441 | 0.364 | -0.739 | 0.188 | 1.173 | -1.406 | 0.534 | -1.167 | -0.494 | -0.254 | -0.739 | -1.279 | 0.102 | 0.387 | 0.216 |