Model info
| Transcription factor | Mef2d | ||||||||
| Model | MEF2D_MOUSE.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 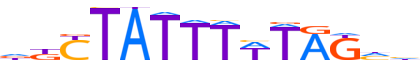 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dKYTATTTWTAKhh | ||||||||
| Best auROC (human) | 0.812 | ||||||||
| Best auROC (mouse) | 0.97 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 27 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Regulators of differentiation {5.1.1} | ||||||||
| TF subfamily | MEF-2 {5.1.1.1} | ||||||||
| MGI | MGI:99533 | ||||||||
| EntrezGene | GeneID:17261 (SSTAR profile) | ||||||||
| UniProt ID | MEF2D_MOUSE | ||||||||
| UniProt AC | Q63943 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Mef2d expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 147.0 | 30.0 | 151.0 | 172.0 |
| 02 | 36.0 | 37.0 | 244.0 | 183.0 |
| 03 | 13.0 | 377.0 | 30.0 | 80.0 |
| 04 | 1.0 | 12.0 | 3.0 | 484.0 |
| 05 | 481.0 | 0.0 | 2.0 | 17.0 |
| 06 | 29.0 | 2.0 | 2.0 | 467.0 |
| 07 | 40.0 | 11.0 | 4.0 | 445.0 |
| 08 | 45.0 | 16.0 | 3.0 | 436.0 |
| 09 | 148.0 | 45.0 | 7.0 | 300.0 |
| 10 | 38.0 | 5.0 | 25.0 | 432.0 |
| 11 | 382.0 | 0.0 | 112.0 | 6.0 |
| 12 | 43.0 | 17.0 | 394.0 | 46.0 |
| 13 | 151.0 | 237.0 | 32.0 | 80.0 |
| 14 | 155.0 | 143.0 | 43.0 | 159.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.16 | -1.389 | 0.187 | 0.316 |
| 02 | -1.215 | -1.189 | 0.663 | 0.377 |
| 03 | -2.163 | 1.096 | -1.389 | -0.439 |
| 04 | -3.903 | -2.234 | -3.325 | 1.345 |
| 05 | 1.338 | -4.4 | -3.573 | -1.92 |
| 06 | -1.421 | -3.573 | -3.573 | 1.309 |
| 07 | -1.114 | -2.311 | -3.126 | 1.261 |
| 08 | -1.0 | -1.975 | -3.325 | 1.241 |
| 09 | 0.167 | -1.0 | -2.694 | 0.868 |
| 10 | -1.163 | -2.961 | -1.561 | 1.231 |
| 11 | 1.109 | -4.4 | -0.108 | -2.819 |
| 12 | -1.044 | -1.92 | 1.14 | -0.979 |
| 13 | 0.187 | 0.634 | -1.328 | -0.439 |
| 14 | 0.213 | 0.133 | -1.044 | 0.238 |