Model info
| Transcription factor | Mybl1 | ||||||||
| Model | MYBA_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 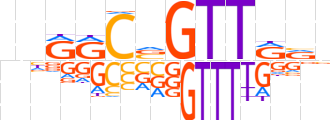 | ||||||||
| LOGO (reverse complement) | 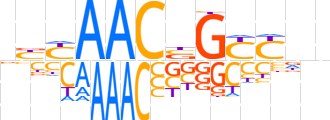 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bnRRCvGTTRRn | ||||||||
| Best auROC (human) | 0.415 | ||||||||
| Best auROC (mouse) | 0.931 | ||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 393 | ||||||||
| TF family | Myb/SANT domain factors {3.5.1} | ||||||||
| TF subfamily | Myb-like factors {3.5.1.1} | ||||||||
| MGI | MGI:99925 | ||||||||
| EntrezGene | GeneID:17864 (SSTAR profile) | ||||||||
| UniProt ID | MYBA_MOUSE | ||||||||
| UniProt AC | P51960 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Mybl1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 16.0 | 22.0 | 11.0 | 8.0 | 12.0 | 29.0 | 17.0 | 38.0 | 38.0 | 74.0 | 24.0 | 35.0 | 4.0 | 18.0 | 10.0 | 26.0 |
| 02 | 16.0 | 1.0 | 50.0 | 3.0 | 35.0 | 1.0 | 93.0 | 14.0 | 19.0 | 2.0 | 38.0 | 3.0 | 10.0 | 3.0 | 90.0 | 4.0 |
| 03 | 12.0 | 0.0 | 67.0 | 1.0 | 1.0 | 0.0 | 5.0 | 1.0 | 60.0 | 4.0 | 186.0 | 21.0 | 6.0 | 1.0 | 11.0 | 6.0 |
| 04 | 2.0 | 74.0 | 3.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 13.0 | 251.0 | 5.0 | 0.0 | 0.0 | 27.0 | 1.0 | 1.0 |
| 05 | 5.0 | 8.0 | 2.0 | 0.0 | 94.0 | 135.0 | 126.0 | 2.0 | 2.0 | 7.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 100.0 | 1.0 | 0.0 | 0.0 | 151.0 | 0.0 | 0.0 | 0.0 | 127.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 380.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 382.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 87.0 | 7.0 | 244.0 | 44.0 |
| 10 | 19.0 | 29.0 | 28.0 | 11.0 | 0.0 | 4.0 | 2.0 | 1.0 | 40.0 | 26.0 | 169.0 | 9.0 | 13.0 | 5.0 | 24.0 | 2.0 |
| 11 | 23.0 | 13.0 | 32.0 | 4.0 | 12.0 | 21.0 | 10.0 | 21.0 | 43.0 | 69.0 | 65.0 | 46.0 | 5.0 | 4.0 | 9.0 | 5.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.393 | -0.08 | -0.757 | -1.063 | -0.673 | 0.192 | -0.333 | 0.459 | 0.459 | 1.121 | 0.005 | 0.378 | -1.713 | -0.277 | -0.849 | 0.084 |
| 02 | -0.393 | -2.872 | 0.731 | -1.973 | 0.378 | -2.872 | 1.348 | -0.523 | -0.224 | -2.325 | 0.459 | -1.973 | -0.849 | -1.973 | 1.316 | -1.713 |
| 03 | -0.673 | -4.178 | 1.022 | -2.872 | -2.872 | -4.178 | -1.507 | -2.872 | 0.912 | -1.713 | 2.039 | -0.126 | -1.336 | -2.872 | -0.757 | -1.336 |
| 04 | -2.325 | 1.121 | -1.973 | -4.178 | -4.178 | -1.507 | -4.178 | -4.178 | -0.595 | 2.339 | -1.507 | -4.178 | -4.178 | 0.121 | -2.872 | -2.872 |
| 05 | -1.507 | -1.063 | -2.325 | -4.178 | 1.359 | 1.72 | 1.651 | -2.325 | -2.325 | -1.191 | -4.178 | -4.178 | -4.178 | -2.872 | -4.178 | -4.178 |
| 06 | -4.178 | -4.178 | 1.421 | -2.872 | -4.178 | -4.178 | 1.831 | -4.178 | -4.178 | -4.178 | 1.659 | -2.872 | -4.178 | -4.178 | -2.325 | -4.178 |
| 07 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | 2.753 | -4.178 | -4.178 | -4.178 | -2.325 |
| 08 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | 2.758 |
| 09 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | -4.178 | 1.282 | -1.191 | 2.31 | 0.604 |
| 10 | -0.224 | 0.192 | 0.157 | -0.757 | -4.178 | -1.713 | -2.325 | -2.872 | 0.51 | 0.084 | 1.944 | -0.951 | -0.595 | -1.507 | 0.005 | -2.325 |
| 11 | -0.037 | -0.595 | 0.289 | -1.713 | -0.673 | -0.126 | -0.849 | -0.126 | 0.582 | 1.051 | 0.992 | 0.648 | -1.507 | -1.713 | -0.951 | -1.507 |