Model info
| Transcription factor | MYB (GeneCards) | ||||||||
| Model | MYB_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 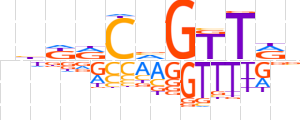 | ||||||||
| LOGO (reverse complement) | 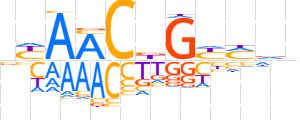 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 11 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nhKRCvGTTRn | ||||||||
| Best auROC (human) | 0.826 | ||||||||
| Best auROC (mouse) | 0.928 | ||||||||
| Peak sets in benchmark (human) | 27 | ||||||||
| Peak sets in benchmark (mouse) | 9 | ||||||||
| Aligned words | 379 | ||||||||
| TF family | Myb/SANT domain factors {3.5.1} | ||||||||
| TF subfamily | Myb-like factors {3.5.1.1} | ||||||||
| HGNC | HGNC:7545 | ||||||||
| EntrezGene | GeneID:4602 (SSTAR profile) | ||||||||
| UniProt ID | MYB_HUMAN | ||||||||
| UniProt AC | P10242 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MYB expression | ||||||||
| ReMap ChIP-seq dataset list | MYB datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 20.0 | 16.0 | 17.0 | 34.0 | 20.0 | 17.0 | 3.0 | 41.0 | 28.0 | 35.0 | 15.0 | 64.0 | 7.0 | 17.0 | 13.0 | 31.0 |
| 02 | 16.0 | 3.0 | 42.0 | 14.0 | 25.0 | 1.0 | 31.0 | 28.0 | 19.0 | 4.0 | 18.0 | 7.0 | 9.0 | 1.0 | 118.0 | 42.0 |
| 03 | 23.0 | 0.0 | 36.0 | 10.0 | 4.0 | 0.0 | 4.0 | 1.0 | 79.0 | 6.0 | 100.0 | 24.0 | 13.0 | 3.0 | 54.0 | 21.0 |
| 04 | 7.0 | 109.0 | 1.0 | 2.0 | 0.0 | 9.0 | 0.0 | 0.0 | 7.0 | 171.0 | 0.0 | 16.0 | 0.0 | 56.0 | 0.0 | 0.0 |
| 05 | 6.0 | 4.0 | 2.0 | 2.0 | 185.0 | 76.0 | 38.0 | 46.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 16.0 | 1.0 |
| 06 | 0.0 | 0.0 | 192.0 | 0.0 | 0.0 | 0.0 | 81.0 | 0.0 | 0.0 | 0.0 | 56.0 | 0.0 | 0.0 | 0.0 | 49.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.0 | 22.0 | 44.0 | 303.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 9.0 | 0.0 | 0.0 | 0.0 | 22.0 | 0.0 | 0.0 | 1.0 | 43.0 | 3.0 | 3.0 | 16.0 | 281.0 |
| 09 | 1.0 | 0.0 | 2.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 11.0 | 1.0 | 104.0 | 2.0 | 187.0 | 62.0 |
| 10 | 32.0 | 24.0 | 41.0 | 16.0 | 2.0 | 0.0 | 0.0 | 0.0 | 39.0 | 53.0 | 73.0 | 35.0 | 19.0 | 8.0 | 21.0 | 15.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.164 | -0.382 | -0.323 | 0.359 | -0.164 | -0.323 | -1.963 | 0.545 | 0.167 | 0.388 | -0.445 | 0.987 | -1.18 | -0.323 | -0.585 | 0.268 |
| 02 | -0.382 | -1.963 | 0.569 | -0.513 | 0.056 | -2.862 | 0.268 | 0.167 | -0.214 | -1.703 | -0.267 | -1.18 | -0.94 | -2.862 | 1.596 | 0.569 |
| 03 | -0.026 | -4.17 | 0.416 | -0.839 | -1.703 | -4.17 | -1.703 | -2.862 | 1.196 | -1.326 | 1.431 | 0.016 | -0.585 | -1.963 | 0.818 | -0.116 |
| 04 | -1.18 | 1.517 | -2.862 | -2.315 | -4.17 | -0.94 | -4.17 | -4.17 | -1.18 | 1.966 | -4.17 | -0.382 | -4.17 | 0.854 | -4.17 | -4.17 |
| 05 | -1.326 | -1.703 | -2.315 | -2.315 | 2.044 | 1.158 | 0.469 | 0.659 | -4.17 | -2.862 | -4.17 | -4.17 | -2.862 | -4.17 | -0.382 | -2.862 |
| 06 | -4.17 | -4.17 | 2.082 | -4.17 | -4.17 | -4.17 | 1.221 | -4.17 | -4.17 | -4.17 | 0.854 | -4.17 | -4.17 | -4.17 | 0.721 | -4.17 |
| 07 | -4.17 | -4.17 | -4.17 | -4.17 | -4.17 | -4.17 | -4.17 | -4.17 | -0.94 | -0.07 | 0.615 | 2.537 | -4.17 | -4.17 | -4.17 | -4.17 |
| 08 | -4.17 | -4.17 | -4.17 | -0.94 | -4.17 | -4.17 | -4.17 | -0.07 | -4.17 | -4.17 | -2.862 | 0.592 | -1.963 | -1.963 | -0.382 | 2.462 |
| 09 | -2.862 | -4.17 | -2.315 | -4.17 | -1.963 | -4.17 | -4.17 | -4.17 | -1.497 | -4.17 | -0.747 | -2.862 | 1.47 | -2.315 | 2.055 | 0.955 |
| 10 | 0.299 | 0.016 | 0.545 | -0.382 | -2.315 | -4.17 | -4.17 | -4.17 | 0.495 | 0.799 | 1.118 | 0.388 | -0.214 | -1.053 | -0.116 | -0.445 |