Model info
| Transcription factor | Myb | ||||||||
| Model | MYB_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 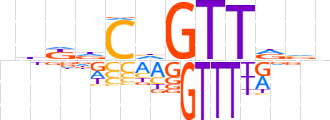 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbddChGTTdvn | ||||||||
| Best auROC (human) | 0.811 | ||||||||
| Best auROC (mouse) | 0.935 | ||||||||
| Peak sets in benchmark (human) | 27 | ||||||||
| Peak sets in benchmark (mouse) | 9 | ||||||||
| Aligned words | 465 | ||||||||
| TF family | Myb/SANT domain factors {3.5.1} | ||||||||
| TF subfamily | Myb-like factors {3.5.1.1} | ||||||||
| MGI | MGI:97249 | ||||||||
| EntrezGene | GeneID:17863 (SSTAR profile) | ||||||||
| UniProt ID | MYB_MOUSE | ||||||||
| UniProt AC | P06876 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Myb expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 26.0 | 13.0 | 35.0 | 37.0 | 14.0 | 19.0 | 8.0 | 53.0 | 24.0 | 48.0 | 24.0 | 61.0 | 12.0 | 9.0 | 24.0 | 51.0 |
| 02 | 23.0 | 1.0 | 40.0 | 12.0 | 20.0 | 7.0 | 31.0 | 31.0 | 22.0 | 7.0 | 43.0 | 19.0 | 24.0 | 5.0 | 137.0 | 36.0 |
| 03 | 28.0 | 4.0 | 47.0 | 10.0 | 12.0 | 4.0 | 2.0 | 2.0 | 78.0 | 16.0 | 108.0 | 49.0 | 15.0 | 5.0 | 34.0 | 44.0 |
| 04 | 6.0 | 120.0 | 7.0 | 0.0 | 1.0 | 28.0 | 0.0 | 0.0 | 14.0 | 167.0 | 8.0 | 2.0 | 2.0 | 101.0 | 0.0 | 2.0 |
| 05 | 10.0 | 7.0 | 5.0 | 1.0 | 196.0 | 91.0 | 49.0 | 80.0 | 11.0 | 3.0 | 1.0 | 0.0 | 1.0 | 2.0 | 1.0 | 0.0 |
| 06 | 0.0 | 0.0 | 216.0 | 2.0 | 0.0 | 0.0 | 102.0 | 1.0 | 0.0 | 0.0 | 56.0 | 0.0 | 0.0 | 0.0 | 79.0 | 2.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 452.0 | 0.0 | 0.0 | 0.0 | 5.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 1.0 | 6.0 | 448.0 |
| 09 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 4.0 | 1.0 | 182.0 | 23.0 | 191.0 | 53.0 |
| 10 | 53.0 | 48.0 | 61.0 | 21.0 | 4.0 | 10.0 | 5.0 | 4.0 | 45.0 | 45.0 | 92.0 | 14.0 | 9.0 | 10.0 | 31.0 | 6.0 |
| 11 | 30.0 | 32.0 | 31.0 | 18.0 | 38.0 | 23.0 | 3.0 | 49.0 | 34.0 | 48.0 | 45.0 | 62.0 | 9.0 | 9.0 | 21.0 | 6.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.095 | -0.774 | 0.199 | 0.254 | -0.702 | -0.403 | -1.241 | 0.61 | -0.174 | 0.512 | -0.174 | 0.75 | -0.851 | -1.129 | -0.174 | 0.572 |
| 02 | -0.216 | -3.043 | 0.331 | -0.851 | -0.353 | -1.368 | 0.079 | 0.079 | -0.259 | -1.368 | 0.402 | -0.403 | -0.174 | -1.684 | 1.555 | 0.227 |
| 03 | -0.022 | -1.89 | 0.491 | -1.027 | -0.851 | -1.89 | -2.499 | -2.499 | 0.994 | -0.571 | 1.318 | 0.532 | -0.634 | -1.684 | 0.17 | 0.425 |
| 04 | -1.514 | 1.423 | -1.368 | -4.327 | -3.043 | -0.022 | -4.327 | -4.327 | -0.702 | 1.753 | -1.241 | -2.499 | -2.499 | 1.251 | -4.327 | -2.499 |
| 05 | -1.027 | -1.368 | -1.684 | -3.043 | 1.912 | 1.147 | 0.532 | 1.019 | -0.935 | -2.149 | -3.043 | -4.327 | -3.043 | -2.499 | -3.043 | -4.327 |
| 06 | -4.327 | -4.327 | 2.009 | -2.499 | -4.327 | -4.327 | 1.261 | -3.043 | -4.327 | -4.327 | 0.665 | -4.327 | -4.327 | -4.327 | 1.007 | -2.499 |
| 07 | -4.327 | -4.327 | -4.327 | -4.327 | -4.327 | -4.327 | -4.327 | -4.327 | -4.327 | -3.043 | -4.327 | 2.747 | -4.327 | -4.327 | -4.327 | -1.684 |
| 08 | -4.327 | -4.327 | -4.327 | -4.327 | -4.327 | -4.327 | -4.327 | -3.043 | -4.327 | -4.327 | -4.327 | -4.327 | -2.499 | -3.043 | -1.514 | 2.738 |
| 09 | -4.327 | -4.327 | -3.043 | -3.043 | -4.327 | -4.327 | -4.327 | -3.043 | -3.043 | -4.327 | -1.89 | -3.043 | 1.839 | -0.216 | 1.887 | 0.61 |
| 10 | 0.61 | 0.512 | 0.75 | -0.305 | -1.89 | -1.027 | -1.684 | -1.89 | 0.448 | 0.448 | 1.158 | -0.702 | -1.129 | -1.027 | 0.079 | -1.514 |
| 11 | 0.046 | 0.11 | 0.079 | -0.456 | 0.28 | -0.216 | -2.149 | 0.532 | 0.17 | 0.512 | 0.448 | 0.766 | -1.129 | -1.129 | -0.305 | -1.514 |