Model info
| Transcription factor | Mycn | ||||||||
| Model | MYCN_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 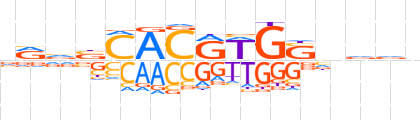 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vRvbMACRWGSnvvv | ||||||||
| Best auROC (human) | 0.921 | ||||||||
| Best auROC (mouse) | 0.961 | ||||||||
| Peak sets in benchmark (human) | 32 | ||||||||
| Peak sets in benchmark (mouse) | 10 | ||||||||
| Aligned words | 493 | ||||||||
| TF family | bHLH-ZIP factors {1.2.6} | ||||||||
| TF subfamily | Myc / Max factors {1.2.6.5} | ||||||||
| MGI | MGI:97357 | ||||||||
| EntrezGene | GeneID:18109 (SSTAR profile) | ||||||||
| UniProt ID | MYCN_MOUSE | ||||||||
| UniProt AC | P03966 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Mycn expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 21.0 | 3.0 | 63.0 | 3.0 | 47.0 | 9.0 | 55.0 | 10.0 | 59.0 | 26.0 | 109.0 | 7.0 | 17.0 | 5.0 | 47.0 | 4.0 |
| 02 | 40.0 | 30.0 | 61.0 | 13.0 | 26.0 | 2.0 | 12.0 | 3.0 | 105.0 | 40.0 | 103.0 | 26.0 | 5.0 | 7.0 | 11.0 | 1.0 |
| 03 | 17.0 | 43.0 | 95.0 | 21.0 | 6.0 | 31.0 | 36.0 | 6.0 | 10.0 | 82.0 | 72.0 | 23.0 | 0.0 | 5.0 | 32.0 | 6.0 |
| 04 | 2.0 | 25.0 | 6.0 | 0.0 | 30.0 | 124.0 | 1.0 | 6.0 | 27.0 | 191.0 | 11.0 | 6.0 | 4.0 | 44.0 | 4.0 | 4.0 |
| 05 | 53.0 | 0.0 | 10.0 | 0.0 | 312.0 | 4.0 | 59.0 | 9.0 | 10.0 | 3.0 | 8.0 | 1.0 | 14.0 | 0.0 | 2.0 | 0.0 |
| 06 | 12.0 | 326.0 | 49.0 | 2.0 | 2.0 | 2.0 | 2.0 | 1.0 | 7.0 | 67.0 | 5.0 | 0.0 | 2.0 | 6.0 | 1.0 | 1.0 |
| 07 | 14.0 | 1.0 | 7.0 | 1.0 | 86.0 | 2.0 | 295.0 | 18.0 | 22.0 | 1.0 | 30.0 | 4.0 | 0.0 | 1.0 | 3.0 | 0.0 |
| 08 | 11.0 | 2.0 | 15.0 | 94.0 | 1.0 | 0.0 | 0.0 | 4.0 | 39.0 | 31.0 | 10.0 | 255.0 | 3.0 | 3.0 | 2.0 | 15.0 |
| 09 | 1.0 | 0.0 | 46.0 | 7.0 | 0.0 | 0.0 | 25.0 | 11.0 | 6.0 | 1.0 | 18.0 | 2.0 | 5.0 | 6.0 | 328.0 | 29.0 |
| 10 | 3.0 | 2.0 | 7.0 | 0.0 | 2.0 | 4.0 | 1.0 | 0.0 | 30.0 | 63.0 | 283.0 | 41.0 | 0.0 | 3.0 | 43.0 | 3.0 |
| 11 | 10.0 | 11.0 | 12.0 | 2.0 | 19.0 | 15.0 | 23.0 | 15.0 | 83.0 | 120.0 | 77.0 | 54.0 | 11.0 | 8.0 | 16.0 | 9.0 |
| 12 | 36.0 | 17.0 | 53.0 | 17.0 | 36.0 | 31.0 | 56.0 | 31.0 | 41.0 | 22.0 | 58.0 | 7.0 | 14.0 | 8.0 | 46.0 | 12.0 |
| 13 | 33.0 | 17.0 | 64.0 | 13.0 | 37.0 | 10.0 | 18.0 | 13.0 | 67.0 | 43.0 | 79.0 | 24.0 | 14.0 | 6.0 | 31.0 | 16.0 |
| 14 | 41.0 | 21.0 | 72.0 | 17.0 | 9.0 | 35.0 | 26.0 | 6.0 | 37.0 | 71.0 | 66.0 | 18.0 | 3.0 | 22.0 | 35.0 | 6.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.361 | -2.204 | 0.725 | -2.204 | 0.434 | -1.185 | 0.59 | -1.084 | 0.66 | -0.151 | 1.271 | -1.425 | -0.569 | -1.74 | 0.434 | -1.946 |
| 02 | 0.274 | -0.01 | 0.693 | -0.83 | -0.151 | -2.554 | -0.908 | -2.204 | 1.233 | 0.274 | 1.214 | -0.151 | -1.74 | -1.425 | -0.992 | -3.097 |
| 03 | -0.569 | 0.346 | 1.134 | -0.361 | -1.57 | 0.022 | 0.17 | -1.57 | -1.084 | 0.987 | 0.858 | -0.272 | -4.375 | -1.74 | 0.054 | -1.57 |
| 04 | -2.554 | -0.19 | -1.57 | -4.375 | -0.01 | 1.399 | -3.097 | -1.57 | -0.114 | 1.83 | -0.992 | -1.57 | -1.946 | 0.369 | -1.946 | -1.946 |
| 05 | 0.553 | -4.375 | -1.084 | -4.375 | 2.32 | -1.946 | 0.66 | -1.185 | -1.084 | -2.204 | -1.298 | -3.097 | -0.758 | -4.375 | -2.554 | -4.375 |
| 06 | -0.908 | 2.364 | 0.475 | -2.554 | -2.554 | -2.554 | -2.554 | -3.097 | -1.425 | 0.786 | -1.74 | -4.375 | -2.554 | -1.57 | -3.097 | -3.097 |
| 07 | -0.758 | -3.097 | -1.425 | -3.097 | 1.035 | -2.554 | 2.264 | -0.513 | -0.316 | -3.097 | -0.01 | -1.946 | -4.375 | -3.097 | -2.204 | -4.375 |
| 08 | -0.992 | -2.554 | -0.691 | 1.123 | -3.097 | -4.375 | -4.375 | -1.946 | 0.249 | 0.022 | -1.084 | 2.119 | -2.204 | -2.204 | -2.554 | -0.691 |
| 09 | -3.097 | -4.375 | 0.413 | -1.425 | -4.375 | -4.375 | -0.19 | -0.992 | -1.57 | -3.097 | -0.513 | -2.554 | -1.74 | -1.57 | 2.37 | -0.044 |
| 10 | -2.204 | -2.554 | -1.425 | -4.375 | -2.554 | -1.946 | -3.097 | -4.375 | -0.01 | 0.725 | 2.223 | 0.299 | -4.375 | -2.204 | 0.346 | -2.204 |
| 11 | -1.084 | -0.992 | -0.908 | -2.554 | -0.46 | -0.691 | -0.272 | -0.691 | 0.999 | 1.366 | 0.925 | 0.572 | -0.992 | -1.298 | -0.628 | -1.185 |
| 12 | 0.17 | -0.569 | 0.553 | -0.569 | 0.17 | 0.022 | 0.608 | 0.022 | 0.299 | -0.316 | 0.643 | -1.425 | -0.758 | -1.298 | 0.413 | -0.908 |
| 13 | 0.084 | -0.569 | 0.741 | -0.83 | 0.197 | -1.084 | -0.513 | -0.83 | 0.786 | 0.346 | 0.95 | -0.23 | -0.758 | -1.57 | 0.022 | -0.628 |
| 14 | 0.299 | -0.361 | 0.858 | -0.569 | -1.185 | 0.142 | -0.151 | -1.57 | 0.197 | 0.844 | 0.771 | -0.513 | -2.204 | -0.316 | 0.142 | -1.57 |