Model info
| Transcription factor | Myod1 | ||||||||
| Model | MYOD1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 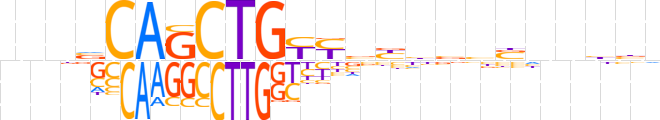 | ||||||||
| LOGO (reverse complement) | 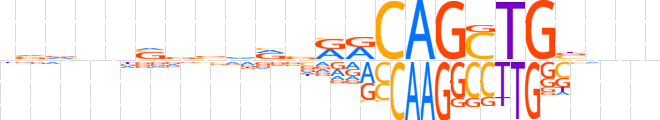 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnnvCAGCTGYYbbhbbbnnbbn | ||||||||
| Best auROC (human) | 0.993 | ||||||||
| Best auROC (mouse) | 0.998 | ||||||||
| Peak sets in benchmark (human) | 9 | ||||||||
| Peak sets in benchmark (mouse) | 63 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | MyoD / ASC-related factors {1.2.2} | ||||||||
| TF subfamily | Myogenic transcription factors {1.2.2.1} | ||||||||
| MGI | MGI:97275 | ||||||||
| EntrezGene | GeneID:17927 (SSTAR profile) | ||||||||
| UniProt ID | MYOD1_MOUSE | ||||||||
| UniProt AC | P10085 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Myod1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 14.388 | 34.858 | 41.409 | 15.562 | 43.362 | 51.969 | 11.077 | 45.118 | 20.098 | 48.35 | 43.474 | 17.435 | 8.733 | 41.809 | 49.097 | 13.261 |
| 02 | 13.524 | 13.103 | 44.081 | 15.872 | 49.745 | 28.035 | 11.395 | 87.811 | 32.867 | 39.353 | 51.046 | 21.792 | 5.959 | 23.436 | 49.097 | 12.884 |
| 03 | 13.91 | 20.286 | 64.95 | 2.951 | 33.774 | 34.489 | 11.294 | 24.37 | 24.487 | 52.019 | 69.973 | 9.14 | 4.838 | 27.172 | 100.817 | 5.531 |
| 04 | 0.0 | 77.009 | 0.0 | 0.0 | 0.0 | 133.966 | 0.0 | 0.0 | 0.92 | 245.011 | 1.102 | 0.0 | 0.0 | 41.991 | 0.0 | 0.0 |
| 05 | 0.92 | 0.0 | 0.0 | 0.0 | 492.926 | 0.939 | 1.012 | 3.099 | 1.102 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 1.144 | 99.903 | 385.039 | 8.863 | 0.0 | 0.939 | 0.0 | 0.0 | 0.0 | 0.0 | 1.012 | 0.0 | 0.0 | 0.0 | 3.099 | 0.0 |
| 07 | 0.0 | 1.144 | 0.0 | 0.0 | 0.912 | 98.903 | 0.0 | 1.027 | 0.0 | 388.246 | 0.0 | 0.904 | 0.0 | 8.863 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.912 | 0.0 | 0.98 | 0.0 | 496.176 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.932 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.98 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.704 | 495.316 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.704 | 0.0 | 0.0 | 2.049 | 214.636 | 19.313 | 260.298 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 0.0 | 0.938 | 1.111 | 0.0 | 2.004 | 67.584 | 3.887 | 144.865 | 0.0 | 10.36 | 1.16 | 7.793 | 0.0 | 149.883 | 25.022 | 85.393 |
| 12 | 0.0 | 2.004 | 0.0 | 0.0 | 52.125 | 101.29 | 16.616 | 58.734 | 0.931 | 21.061 | 9.188 | 0.0 | 11.765 | 84.4 | 120.594 | 21.293 |
| 13 | 4.89 | 34.996 | 17.858 | 7.077 | 9.83 | 123.601 | 13.031 | 62.292 | 14.114 | 69.398 | 32.517 | 30.37 | 2.939 | 46.975 | 9.812 | 20.301 |
| 14 | 3.138 | 10.829 | 13.01 | 4.795 | 56.817 | 76.572 | 1.994 | 139.587 | 11.532 | 28.907 | 19.171 | 13.608 | 8.895 | 48.893 | 37.26 | 24.991 |
| 15 | 7.234 | 15.821 | 48.596 | 8.731 | 47.203 | 64.367 | 14.031 | 39.601 | 8.657 | 25.526 | 29.323 | 7.929 | 2.782 | 27.535 | 134.26 | 18.404 |
| 16 | 3.92 | 15.919 | 31.964 | 14.073 | 35.755 | 50.712 | 9.614 | 37.168 | 21.708 | 111.693 | 34.754 | 58.056 | 1.045 | 29.572 | 32.434 | 11.613 |
| 17 | 2.965 | 20.797 | 30.674 | 7.992 | 17.021 | 121.262 | 7.467 | 62.145 | 7.754 | 62.113 | 26.465 | 12.436 | 2.841 | 81.436 | 28.627 | 8.007 |
| 18 | 7.101 | 7.881 | 11.757 | 3.842 | 99.772 | 89.823 | 9.199 | 86.813 | 7.643 | 36.053 | 36.439 | 13.099 | 5.247 | 45.18 | 27.741 | 12.411 |
| 19 | 17.743 | 43.792 | 48.182 | 10.045 | 60.628 | 55.164 | 11.258 | 51.886 | 14.855 | 33.462 | 21.046 | 15.774 | 8.974 | 29.522 | 63.123 | 14.546 |
| 20 | 10.669 | 14.703 | 26.36 | 50.468 | 28.619 | 38.646 | 7.785 | 86.89 | 19.511 | 43.026 | 23.472 | 57.601 | 8.675 | 15.038 | 36.492 | 32.045 |
| 21 | 8.466 | 20.229 | 28.138 | 10.64 | 26.016 | 37.113 | 5.918 | 42.367 | 11.789 | 29.842 | 28.208 | 24.27 | 7.939 | 140.614 | 47.701 | 30.75 |
| 22 | 6.027 | 15.859 | 23.533 | 8.791 | 102.931 | 49.195 | 12.661 | 63.01 | 17.402 | 36.984 | 41.074 | 14.504 | 8.568 | 41.929 | 39.059 | 18.471 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.761 | 0.108 | 0.278 | -0.685 | 0.324 | 0.504 | -1.015 | 0.363 | -0.435 | 0.432 | 0.327 | -0.574 | -1.244 | 0.288 | 0.447 | -0.841 |
| 02 | -0.822 | -0.852 | 0.34 | -0.666 | 0.46 | -0.107 | -0.988 | 1.025 | 0.05 | 0.228 | 0.486 | -0.355 | -1.606 | -0.284 | 0.447 | -0.869 |
| 03 | -0.794 | -0.425 | 0.725 | -2.249 | 0.077 | 0.097 | -0.996 | -0.245 | -0.24 | 0.505 | 0.799 | -1.2 | -1.801 | -0.138 | 1.163 | -1.676 |
| 04 | -4.4 | 0.895 | -4.4 | -4.4 | -4.4 | 1.446 | -4.4 | -4.4 | -3.185 | 2.049 | -3.055 | -4.4 | -4.4 | 0.292 | -4.4 | -4.4 |
| 05 | -3.185 | -4.4 | -4.4 | -4.4 | 2.747 | -3.171 | -3.118 | -2.205 | -3.055 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 06 | -3.027 | 1.154 | 2.5 | -1.23 | -4.4 | -3.171 | -4.4 | -4.4 | -4.4 | -4.4 | -3.118 | -4.4 | -4.4 | -4.4 | -2.205 | -4.4 |
| 07 | -4.4 | -3.027 | -4.4 | -4.4 | -3.192 | 1.144 | -4.4 | -3.107 | -4.4 | 2.508 | -4.4 | -3.198 | -4.4 | -1.23 | -4.4 | -4.4 |
| 08 | -4.4 | -4.4 | -4.4 | -3.192 | -4.4 | -3.141 | -4.4 | 2.753 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.613 |
| 09 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.141 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.045 | 2.752 | -4.4 |
| 10 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.045 | -4.4 | -4.4 | -2.563 | 1.916 | -0.474 | 2.109 | -4.4 | -4.4 | -4.4 | -4.4 |
| 11 | -4.4 | -3.172 | -3.049 | -4.4 | -2.582 | 0.765 | -2.001 | 1.524 | -4.4 | -1.08 | -3.017 | -1.352 | -4.4 | 1.558 | -0.219 | 0.997 |
| 12 | -4.4 | -2.582 | -4.4 | -4.4 | 0.507 | 1.167 | -0.621 | 0.625 | -3.177 | -0.389 | -1.195 | -4.4 | -0.957 | 0.986 | 1.341 | -0.378 |
| 13 | -1.791 | 0.112 | -0.55 | -1.444 | -1.13 | 1.366 | -0.858 | 0.684 | -0.78 | 0.791 | 0.039 | -0.028 | -2.252 | 0.403 | -1.132 | -0.425 |
| 14 | -2.194 | -1.037 | -0.859 | -1.809 | 0.592 | 0.889 | -2.586 | 1.487 | -0.976 | -0.077 | -0.481 | -0.816 | -1.226 | 0.443 | 0.174 | -0.22 |
| 15 | -1.423 | -0.669 | 0.437 | -1.244 | 0.408 | 0.716 | -0.786 | 0.234 | -1.252 | -0.2 | -0.063 | -1.336 | -2.3 | -0.125 | 1.448 | -0.521 |
| 16 | -1.994 | -0.663 | 0.022 | -0.783 | 0.133 | 0.479 | -1.152 | 0.171 | -0.359 | 1.265 | 0.105 | 0.614 | -3.094 | -0.055 | 0.037 | -0.969 |
| 17 | -2.244 | -0.401 | -0.018 | -1.329 | -0.597 | 1.347 | -1.393 | 0.681 | -1.357 | 0.681 | -0.164 | -0.903 | -2.282 | 0.95 | -0.087 | -1.327 |
| 18 | -1.441 | -1.342 | -0.957 | -2.012 | 1.152 | 1.048 | -1.194 | 1.014 | -1.371 | 0.141 | 0.152 | -0.853 | -1.725 | 0.365 | -0.118 | -0.905 |
| 19 | -0.557 | 0.334 | 0.429 | -1.109 | 0.657 | 0.563 | -0.999 | 0.502 | -0.73 | 0.068 | -0.389 | -0.672 | -1.218 | -0.056 | 0.697 | -0.751 |
| 20 | -1.051 | -0.74 | -0.168 | 0.475 | -0.087 | 0.21 | -1.354 | 1.015 | -0.464 | 0.316 | -0.282 | 0.606 | -1.25 | -0.718 | 0.153 | 0.025 |
| 21 | -1.273 | -0.428 | -0.104 | -1.054 | -0.181 | 0.17 | -1.613 | 0.301 | -0.955 | -0.046 | -0.101 | -0.249 | -1.335 | 1.494 | 0.419 | -0.016 |
| 22 | -1.596 | -0.666 | -0.28 | -1.237 | 1.183 | 0.449 | -0.886 | 0.695 | -0.576 | 0.167 | 0.27 | -0.753 | -1.262 | 0.291 | 0.221 | -0.517 |