Model info
| Transcription factor | Myog | ||||||||
| Model | MYOG_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 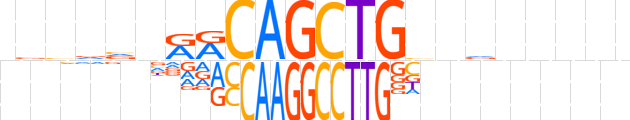 | ||||||||
| LOGO (reverse complement) | 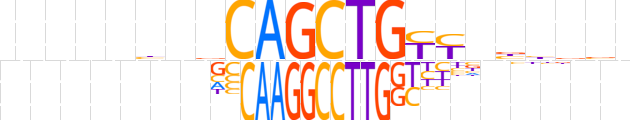 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvvvvnRRCAGCTGbndnnnnn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.997 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 20 | ||||||||
| Aligned words | 461 | ||||||||
| TF family | MyoD / ASC-related factors {1.2.2} | ||||||||
| TF subfamily | Myogenic transcription factors {1.2.2.1} | ||||||||
| MGI | MGI:97276 | ||||||||
| EntrezGene | GeneID:17928 (SSTAR profile) | ||||||||
| UniProt ID | MYOG_MOUSE | ||||||||
| UniProt AC | P12979 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Myog expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 18.724 | 25.863 | 58.434 | 8.612 | 31.737 | 38.484 | 10.979 | 30.525 | 25.75 | 51.594 | 52.295 | 14.323 | 8.925 | 33.955 | 42.959 | 5.021 |
| 02 | 20.342 | 22.901 | 39.225 | 2.668 | 61.636 | 53.683 | 4.893 | 29.684 | 37.004 | 60.746 | 58.122 | 8.795 | 7.259 | 30.076 | 16.3 | 4.846 |
| 03 | 37.399 | 18.57 | 67.188 | 3.085 | 86.358 | 44.058 | 4.884 | 32.106 | 46.077 | 22.494 | 45.142 | 4.829 | 4.106 | 10.071 | 28.088 | 3.726 |
| 04 | 38.147 | 32.905 | 95.046 | 7.843 | 48.0 | 31.425 | 7.045 | 8.723 | 41.534 | 35.869 | 59.996 | 7.903 | 9.364 | 5.708 | 27.669 | 1.005 |
| 05 | 29.372 | 21.3 | 69.405 | 16.968 | 38.518 | 17.426 | 14.915 | 35.047 | 39.991 | 45.012 | 74.196 | 30.556 | 4.83 | 4.987 | 12.762 | 2.894 |
| 06 | 40.735 | 0.0 | 71.976 | 0.0 | 79.611 | 1.967 | 7.147 | 0.0 | 97.188 | 5.808 | 66.275 | 2.008 | 20.813 | 0.0 | 64.652 | 0.0 |
| 07 | 108.848 | 0.0 | 128.432 | 1.067 | 7.775 | 0.0 | 0.0 | 0.0 | 141.126 | 0.0 | 65.162 | 3.762 | 0.0 | 0.0 | 2.008 | 0.0 |
| 08 | 0.0 | 257.749 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 195.602 | 0.0 | 0.0 | 0.0 | 4.829 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 458.18 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 1.04 | 0.0 | 457.14 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 0.0 | 1.04 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 452.013 | 5.127 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 453.053 | 0.0 | 0.0 | 0.0 | 5.127 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 458.18 | 0.0 |
| 14 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 67.109 | 186.688 | 102.752 | 101.631 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 10.505 | 14.396 | 39.427 | 2.781 | 81.334 | 30.927 | 14.309 | 60.118 | 12.506 | 20.664 | 39.982 | 29.599 | 7.789 | 13.957 | 52.897 | 26.988 |
| 16 | 13.357 | 13.141 | 70.106 | 15.529 | 38.683 | 8.018 | 10.782 | 22.461 | 35.958 | 32.939 | 57.498 | 20.221 | 17.19 | 15.645 | 70.217 | 16.434 |
| 17 | 23.718 | 30.919 | 40.06 | 10.491 | 20.28 | 30.021 | 2.969 | 16.474 | 38.568 | 75.62 | 63.022 | 31.393 | 13.636 | 19.128 | 26.909 | 14.971 |
| 18 | 19.263 | 17.678 | 41.256 | 18.005 | 36.702 | 50.038 | 7.935 | 61.014 | 16.306 | 48.091 | 47.824 | 20.74 | 10.443 | 22.42 | 24.785 | 15.682 |
| 19 | 18.009 | 22.463 | 25.449 | 16.791 | 56.382 | 37.282 | 3.749 | 40.814 | 29.886 | 28.715 | 39.913 | 23.285 | 15.142 | 37.734 | 49.335 | 13.23 |
| 20 | 16.484 | 24.073 | 59.097 | 19.766 | 44.942 | 33.609 | 8.804 | 38.839 | 25.429 | 33.062 | 43.19 | 16.765 | 4.918 | 19.13 | 52.826 | 17.247 |
| 21 | 31.686 | 12.695 | 34.562 | 12.828 | 41.356 | 30.465 | 8.813 | 29.239 | 23.123 | 41.991 | 62.45 | 36.355 | 9.777 | 22.05 | 44.807 | 15.983 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.418 | -0.1 | 0.706 | -1.171 | 0.102 | 0.292 | -0.938 | 0.063 | -0.105 | 0.583 | 0.596 | -0.68 | -1.137 | 0.168 | 0.401 | -1.681 |
| 02 | -0.337 | -0.22 | 0.311 | -2.253 | 0.759 | 0.622 | -1.705 | 0.035 | 0.253 | 0.745 | 0.701 | -1.151 | -1.334 | 0.048 | -0.554 | -1.714 |
| 03 | 0.264 | -0.426 | 0.845 | -2.125 | 1.095 | 0.426 | -1.707 | 0.113 | 0.471 | -0.238 | 0.45 | -1.717 | -1.866 | -1.021 | -0.019 | -1.955 |
| 04 | 0.283 | 0.137 | 1.19 | -1.261 | 0.511 | 0.092 | -1.363 | -1.159 | 0.368 | 0.223 | 0.733 | -1.253 | -1.091 | -1.561 | -0.034 | -3.04 |
| 05 | 0.025 | -0.291 | 0.878 | -0.514 | 0.293 | -0.488 | -0.64 | 0.2 | 0.33 | 0.447 | 0.944 | 0.064 | -1.717 | -1.687 | -0.792 | -2.181 |
| 06 | 0.348 | -4.328 | 0.914 | -4.328 | 1.014 | -2.514 | -1.349 | -4.328 | 1.213 | -1.545 | 0.832 | -2.496 | -0.314 | -4.328 | 0.807 | -4.328 |
| 07 | 1.326 | -4.328 | 1.49 | -2.996 | -1.269 | -4.328 | -4.328 | -4.328 | 1.584 | -4.328 | 0.815 | -1.946 | -4.328 | -4.328 | -2.496 | -4.328 |
| 08 | -4.328 | 2.186 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | 1.91 | -4.328 | -4.328 | -4.328 | -1.717 | -4.328 | -4.328 |
| 09 | -4.328 | -4.328 | -4.328 | -4.328 | 2.76 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 |
| 10 | -3.015 | -4.328 | 2.758 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 |
| 11 | -4.328 | -3.015 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | 2.747 | -1.661 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 |
| 12 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | 2.749 | -4.328 | -4.328 | -4.328 | -1.661 | -4.328 | -4.328 | -4.328 | -4.328 |
| 13 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | 2.76 | -4.328 |
| 14 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | -4.328 | 0.844 | 1.864 | 1.268 | 1.257 | -4.328 | -4.328 | -4.328 | -4.328 |
| 15 | -0.98 | -0.675 | 0.316 | -2.216 | 1.035 | 0.076 | -0.681 | 0.735 | -0.812 | -0.321 | 0.33 | 0.033 | -1.267 | -0.705 | 0.608 | -0.058 |
| 16 | -0.748 | -0.763 | 0.888 | -0.601 | 0.297 | -1.24 | -0.955 | -0.239 | 0.225 | 0.138 | 0.69 | -0.342 | -0.502 | -0.594 | 0.889 | -0.546 |
| 17 | -0.186 | 0.076 | 0.332 | -0.982 | -0.34 | 0.047 | -2.158 | -0.543 | 0.294 | 0.963 | 0.782 | 0.091 | -0.728 | -0.397 | -0.061 | -0.637 |
| 18 | -0.39 | -0.474 | 0.361 | -0.456 | 0.245 | 0.552 | -1.25 | 0.749 | -0.553 | 0.513 | 0.508 | -0.318 | -0.986 | -0.241 | -0.142 | -0.591 |
| 19 | -0.456 | -0.239 | -0.116 | -0.525 | 0.671 | 0.261 | -1.949 | 0.35 | 0.042 | 0.003 | 0.328 | -0.204 | -0.626 | 0.273 | 0.538 | -0.757 |
| 20 | -0.543 | -0.171 | 0.718 | -0.365 | 0.446 | 0.158 | -1.15 | 0.301 | -0.117 | 0.142 | 0.406 | -0.526 | -1.7 | -0.397 | 0.606 | -0.498 |
| 21 | 0.1 | -0.797 | 0.186 | -0.787 | 0.363 | 0.061 | -1.149 | 0.021 | -0.211 | 0.379 | 0.773 | 0.236 | -1.05 | -0.257 | 0.443 | -0.573 |