Model info
| Transcription factor | NANOG (GeneCards) | ||||||||
| Model | NANOG_HUMAN.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 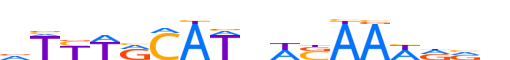 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbhWTTSWnATGYWRWb | ||||||||
| Best auROC (human) | 0.92 | ||||||||
| Best auROC (mouse) | 0.928 | ||||||||
| Peak sets in benchmark (human) | 15 | ||||||||
| Peak sets in benchmark (mouse) | 38 | ||||||||
| Aligned words | 499 | ||||||||
| TF family | NK-related factors {3.1.2} | ||||||||
| TF subfamily | NANOG {3.1.2.12} | ||||||||
| HGNC | HGNC:20857 | ||||||||
| EntrezGene | GeneID:79923 (SSTAR profile) | ||||||||
| UniProt ID | NANOG_HUMAN | ||||||||
| UniProt AC | Q9H9S0 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | NANOG expression | ||||||||
| ReMap ChIP-seq dataset list | NANOG datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 64.0 | 129.0 | 140.0 | 166.0 |
| 02 | 22.0 | 243.0 | 89.0 | 145.0 |
| 03 | 44.0 | 255.0 | 35.0 | 165.0 |
| 04 | 153.0 | 22.0 | 30.0 | 294.0 |
| 05 | 6.0 | 30.0 | 21.0 | 442.0 |
| 06 | 34.0 | 4.0 | 19.0 | 442.0 |
| 07 | 55.0 | 124.0 | 294.0 | 26.0 |
| 08 | 188.0 | 21.0 | 18.0 | 272.0 |
| 09 | 115.0 | 130.0 | 96.0 | 158.0 |
| 10 | 403.0 | 17.0 | 20.0 | 59.0 |
| 11 | 34.0 | 7.0 | 15.0 | 443.0 |
| 12 | 13.0 | 9.0 | 405.0 | 72.0 |
| 13 | 14.0 | 317.0 | 58.0 | 110.0 |
| 14 | 340.0 | 16.0 | 8.0 | 135.0 |
| 15 | 331.0 | 25.0 | 120.0 | 23.0 |
| 16 | 402.0 | 24.0 | 34.0 | 39.0 |
| 17 | 70.0 | 75.0 | 100.0 | 254.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.656 | 0.033 | 0.114 | 0.283 |
| 02 | -1.679 | 0.661 | -0.333 | 0.149 |
| 03 | -1.02 | 0.709 | -1.24 | 0.277 |
| 04 | 0.202 | -1.679 | -1.387 | 0.85 |
| 05 | -2.817 | -1.387 | -1.723 | 1.256 |
| 06 | -1.268 | -3.124 | -1.816 | 1.256 |
| 07 | -0.804 | -0.006 | 0.85 | -1.523 |
| 08 | 0.406 | -1.723 | -1.866 | 0.773 |
| 09 | -0.08 | 0.041 | -0.258 | 0.234 |
| 10 | 1.164 | -1.918 | -1.768 | -0.735 |
| 11 | -1.268 | -2.692 | -2.032 | 1.258 |
| 12 | -2.161 | -2.482 | 1.169 | -0.541 |
| 13 | -2.094 | 0.925 | -0.752 | -0.124 |
| 14 | 0.995 | -1.973 | -2.582 | 0.078 |
| 15 | 0.968 | -1.56 | -0.038 | -1.638 |
| 16 | 1.162 | -1.598 | -1.268 | -1.136 |
| 17 | -0.568 | -0.501 | -0.218 | 0.705 |