Model info
| Transcription factor | Nanog | ||||||||
| Model | NANOG_MOUSE.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 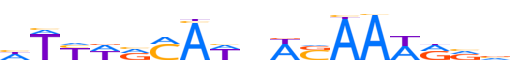 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bbYWTTSWnWTKYWRAd | ||||||||
| Best auROC (human) | 0.914 | ||||||||
| Best auROC (mouse) | 0.939 | ||||||||
| Peak sets in benchmark (human) | 15 | ||||||||
| Peak sets in benchmark (mouse) | 38 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | NK-related factors {3.1.2} | ||||||||
| TF subfamily | NANOG {3.1.2.12} | ||||||||
| MGI | MGI:1919200 | ||||||||
| EntrezGene | GeneID:71950 (SSTAR profile) | ||||||||
| UniProt ID | NANOG_MOUSE | ||||||||
| UniProt AC | Q80Z64 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nanog expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 51.0 | 118.0 | 125.0 | 206.0 |
| 02 | 21.0 | 267.0 | 88.0 | 124.0 |
| 03 | 33.0 | 265.0 | 15.0 | 187.0 |
| 04 | 197.0 | 9.0 | 9.0 | 285.0 |
| 05 | 6.0 | 10.0 | 14.0 | 470.0 |
| 06 | 19.0 | 4.0 | 19.0 | 458.0 |
| 07 | 26.0 | 132.0 | 302.0 | 40.0 |
| 08 | 114.0 | 23.0 | 19.0 | 344.0 |
| 09 | 85.0 | 130.0 | 103.0 | 182.0 |
| 10 | 311.0 | 25.0 | 41.0 | 123.0 |
| 11 | 20.0 | 11.0 | 22.0 | 447.0 |
| 12 | 11.0 | 13.0 | 325.0 | 151.0 |
| 13 | 23.0 | 271.0 | 61.0 | 145.0 |
| 14 | 272.0 | 31.0 | 17.0 | 180.0 |
| 15 | 332.0 | 25.0 | 92.0 | 51.0 |
| 16 | 406.0 | 12.0 | 26.0 | 56.0 |
| 17 | 98.0 | 43.0 | 77.0 | 282.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.879 | -0.057 | 0.0 | 0.495 |
| 02 | -1.725 | 0.752 | -0.346 | -0.008 |
| 03 | -1.298 | 0.745 | -2.034 | 0.399 |
| 04 | 0.45 | -2.484 | -2.484 | 0.817 |
| 05 | -2.819 | -2.394 | -2.096 | 1.315 |
| 06 | -1.818 | -3.126 | -1.818 | 1.29 |
| 07 | -1.525 | 0.054 | 0.875 | -1.114 |
| 08 | -0.091 | -1.64 | -1.818 | 1.004 |
| 09 | -0.38 | 0.039 | -0.191 | 0.372 |
| 10 | 0.904 | -1.561 | -1.09 | -0.016 |
| 11 | -1.77 | -2.311 | -1.681 | 1.265 |
| 12 | -2.311 | -2.163 | 0.948 | 0.187 |
| 13 | -1.64 | 0.767 | -0.705 | 0.147 |
| 14 | 0.771 | -1.358 | -1.92 | 0.361 |
| 15 | 0.969 | -1.561 | -0.302 | -0.879 |
| 16 | 1.17 | -2.234 | -1.525 | -0.788 |
| 17 | -0.24 | -1.044 | -0.477 | 0.807 |