Model info
| Transcription factor | NFATC1 (GeneCards) | ||||||||
| Model | NFAC1_HUMAN.H11MO.0.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 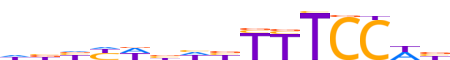 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vWGGAAAvdvWRvvd | ||||||||
| Best auROC (human) | 0.868 | ||||||||
| Best auROC (mouse) | 0.795 | ||||||||
| Peak sets in benchmark (human) | 7 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | NFAT-related factors {6.1.3} | ||||||||
| TF subfamily | NFATc1 {6.1.3.0.1} | ||||||||
| HGNC | HGNC:7775 | ||||||||
| EntrezGene | GeneID:4772 (SSTAR profile) | ||||||||
| UniProt ID | NFAC1_HUMAN | ||||||||
| UniProt AC | O95644 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | NFATC1 expression | ||||||||
| ReMap ChIP-seq dataset list | NFATC1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 241.0 | 115.0 | 117.0 | 27.0 |
| 02 | 197.0 | 25.0 | 30.0 | 248.0 |
| 03 | 42.0 | 4.0 | 452.0 | 2.0 |
| 04 | 7.0 | 23.0 | 465.0 | 5.0 |
| 05 | 473.0 | 6.0 | 18.0 | 3.0 |
| 06 | 405.0 | 34.0 | 52.0 | 9.0 |
| 07 | 411.0 | 34.0 | 42.0 | 13.0 |
| 08 | 250.0 | 83.0 | 134.0 | 33.0 |
| 09 | 247.0 | 26.0 | 71.0 | 156.0 |
| 10 | 178.0 | 119.0 | 173.0 | 30.0 |
| 11 | 244.0 | 44.0 | 22.0 | 190.0 |
| 12 | 132.0 | 15.0 | 274.0 | 79.0 |
| 13 | 269.0 | 61.0 | 111.0 | 59.0 |
| 14 | 215.0 | 159.0 | 94.0 | 32.0 |
| 15 | 225.0 | 60.0 | 69.0 | 146.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.651 | -0.082 | -0.065 | -1.489 |
| 02 | 0.45 | -1.561 | -1.389 | 0.679 |
| 03 | -1.067 | -3.126 | 1.276 | -3.573 |
| 04 | -2.694 | -1.64 | 1.305 | -2.961 |
| 05 | 1.322 | -2.819 | -1.868 | -3.325 |
| 06 | 1.167 | -1.27 | -0.86 | -2.484 |
| 07 | 1.182 | -1.27 | -1.067 | -2.163 |
| 08 | 0.687 | -0.403 | 0.069 | -1.298 |
| 09 | 0.675 | -1.525 | -0.556 | 0.219 |
| 10 | 0.35 | -0.049 | 0.322 | -1.389 |
| 11 | 0.663 | -1.022 | -1.681 | 0.415 |
| 12 | 0.054 | -2.034 | 0.778 | -0.452 |
| 13 | 0.76 | -0.705 | -0.117 | -0.737 |
| 14 | 0.537 | 0.238 | -0.281 | -1.328 |
| 15 | 0.582 | -0.721 | -0.584 | 0.154 |