Model info
| Transcription factor | Nfe2 | ||||||||
| Model | NFE2_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 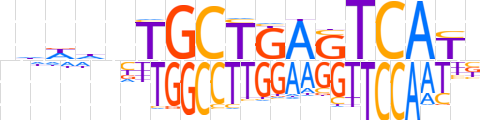 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vRTGACTCAGCAndhhn | ||||||||
| Best auROC (human) | 0.904 | ||||||||
| Best auROC (mouse) | 0.996 | ||||||||
| Peak sets in benchmark (human) | 17 | ||||||||
| Peak sets in benchmark (mouse) | 9 | ||||||||
| Aligned words | 392 | ||||||||
| TF family | Jun-related factors {1.1.1} | ||||||||
| TF subfamily | NF-E2-like factors {1.1.1.2} | ||||||||
| MGI | MGI:97308 | ||||||||
| EntrezGene | GeneID:18022 (SSTAR profile) | ||||||||
| UniProt ID | NFE2_MOUSE | ||||||||
| UniProt AC | Q07279 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nfe2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 57.0 | 0.0 | 36.0 | 1.0 | 94.0 | 4.0 | 4.0 | 0.0 | 110.0 | 5.0 | 33.0 | 0.0 | 15.0 | 4.0 | 20.0 | 0.0 |
| 02 | 1.0 | 0.0 | 1.0 | 274.0 | 0.0 | 0.0 | 0.0 | 13.0 | 0.0 | 0.0 | 0.0 | 93.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 03 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 380.0 | 1.0 |
| 04 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 381.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 05 | 14.0 | 301.0 | 58.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 1.0 | 13.0 | 19.0 | 7.0 | 6.0 | 269.0 | 0.0 | 1.0 | 0.0 | 57.0 | 1.0 | 1.0 | 2.0 | 6.0 |
| 07 | 4.0 | 13.0 | 2.0 | 1.0 | 1.0 | 6.0 | 1.0 | 1.0 | 0.0 | 7.0 | 2.0 | 0.0 | 16.0 | 304.0 | 12.0 | 13.0 |
| 08 | 15.0 | 0.0 | 6.0 | 0.0 | 317.0 | 1.0 | 7.0 | 5.0 | 1.0 | 0.0 | 11.0 | 5.0 | 4.0 | 0.0 | 10.0 | 1.0 |
| 09 | 0.0 | 2.0 | 332.0 | 3.0 | 0.0 | 0.0 | 1.0 | 0.0 | 3.0 | 0.0 | 30.0 | 1.0 | 0.0 | 0.0 | 11.0 | 0.0 |
| 10 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 1.0 | 368.0 | 3.0 | 2.0 | 1.0 | 2.0 | 0.0 | 1.0 |
| 11 | 2.0 | 0.0 | 0.0 | 0.0 | 341.0 | 5.0 | 18.0 | 11.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 2.0 | 1.0 | 0.0 |
| 12 | 80.0 | 64.0 | 141.0 | 59.0 | 5.0 | 2.0 | 0.0 | 1.0 | 3.0 | 6.0 | 5.0 | 6.0 | 3.0 | 1.0 | 5.0 | 2.0 |
| 13 | 38.0 | 6.0 | 18.0 | 29.0 | 23.0 | 8.0 | 3.0 | 39.0 | 45.0 | 14.0 | 23.0 | 69.0 | 8.0 | 13.0 | 13.0 | 34.0 |
| 14 | 63.0 | 11.0 | 13.0 | 27.0 | 17.0 | 6.0 | 1.0 | 17.0 | 31.0 | 5.0 | 3.0 | 18.0 | 15.0 | 19.0 | 7.0 | 130.0 |
| 15 | 54.0 | 20.0 | 28.0 | 24.0 | 14.0 | 10.0 | 1.0 | 16.0 | 6.0 | 5.0 | 4.0 | 9.0 | 58.0 | 40.0 | 15.0 | 79.0 |
| 16 | 70.0 | 18.0 | 24.0 | 20.0 | 18.0 | 20.0 | 3.0 | 34.0 | 11.0 | 12.0 | 12.0 | 13.0 | 20.0 | 40.0 | 20.0 | 48.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.859 | -4.18 | 0.403 | -2.875 | 1.356 | -1.716 | -1.716 | -4.18 | 1.513 | -1.51 | 0.317 | -4.18 | -0.458 | -1.716 | -0.177 | -4.18 |
| 02 | -2.875 | -4.18 | -2.875 | 2.424 | -4.18 | -4.18 | -4.18 | -0.598 | -4.18 | -4.18 | -4.18 | 1.346 | -4.18 | -4.18 | -4.18 | -2.875 |
| 03 | -4.18 | -4.18 | -2.875 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -2.875 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | 2.75 | -2.875 |
| 04 | -2.875 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | 2.753 | -4.18 | -4.18 | -4.18 | -2.875 | -4.18 | -4.18 | -4.18 |
| 05 | -0.526 | 2.517 | 0.876 | -0.852 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 | -4.18 |
| 06 | -4.18 | -4.18 | -2.875 | -0.598 | -0.227 | -1.193 | -1.339 | 2.405 | -4.18 | -2.875 | -4.18 | 0.859 | -2.875 | -2.875 | -2.327 | -1.339 |
| 07 | -1.716 | -0.598 | -2.327 | -2.875 | -2.875 | -1.339 | -2.875 | -2.875 | -4.18 | -1.193 | -2.327 | -4.18 | -0.395 | 2.527 | -0.675 | -0.598 |
| 08 | -0.458 | -4.18 | -1.339 | -4.18 | 2.569 | -2.875 | -1.193 | -1.51 | -2.875 | -4.18 | -0.76 | -1.51 | -1.716 | -4.18 | -0.852 | -2.875 |
| 09 | -4.18 | -2.327 | 2.615 | -1.975 | -4.18 | -4.18 | -2.875 | -4.18 | -1.975 | -4.18 | 0.223 | -2.875 | -4.18 | -4.18 | -0.76 | -4.18 |
| 10 | -4.18 | -1.975 | -4.18 | -4.18 | -4.18 | -2.327 | -4.18 | -4.18 | -2.875 | 2.718 | -1.975 | -2.327 | -2.875 | -2.327 | -4.18 | -2.875 |
| 11 | -2.327 | -4.18 | -4.18 | -4.18 | 2.642 | -1.51 | -0.28 | -0.76 | -2.875 | -2.875 | -2.875 | -4.18 | -4.18 | -2.327 | -2.875 | -4.18 |
| 12 | 1.196 | 0.974 | 1.761 | 0.893 | -1.51 | -2.327 | -4.18 | -2.875 | -1.975 | -1.339 | -1.51 | -1.339 | -1.975 | -2.875 | -1.51 | -2.327 |
| 13 | 0.456 | -1.339 | -0.28 | 0.189 | -0.039 | -1.066 | -1.975 | 0.482 | 0.624 | -0.526 | -0.039 | 1.049 | -1.066 | -0.598 | -0.598 | 0.346 |
| 14 | 0.958 | -0.76 | -0.598 | 0.119 | -0.336 | -1.339 | -2.875 | -0.336 | 0.255 | -1.51 | -1.975 | -0.28 | -0.458 | -0.227 | -1.193 | 1.68 |
| 15 | 0.805 | -0.177 | 0.155 | 0.003 | -0.526 | -0.852 | -2.875 | -0.395 | -1.339 | -1.51 | -1.716 | -0.953 | 0.876 | 0.507 | -0.458 | 1.183 |
| 16 | 1.063 | -0.28 | 0.003 | -0.177 | -0.28 | -0.177 | -1.975 | 0.346 | -0.76 | -0.675 | -0.675 | -0.598 | -0.177 | 0.507 | -0.177 | 0.688 |