Model info
| Transcription factor | Nfic | ||||||||
| Model | NFIC_MOUSE.H11MO.1.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 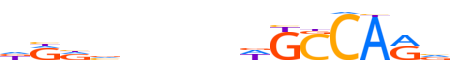 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 1 | ||||||||
| Consensus | bYTGGCWbnnnbhYh | ||||||||
| Best auROC (human) | 0.958 | ||||||||
| Best auROC (mouse) | 0.807 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 1 | ||||||||
| Aligned words | 410 | ||||||||
| TF family | Nuclear factor 1 {7.1.2} | ||||||||
| TF subfamily | NF-1C {7.1.2.0.3} | ||||||||
| MGI | MGI:109591 | ||||||||
| EntrezGene | GeneID:18029 (SSTAR profile) | ||||||||
| UniProt ID | NFIC_MOUSE | ||||||||
| UniProt AC | P70255 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nfic expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 38.0 | 195.0 | 78.0 | 99.0 |
| 02 | 9.0 | 228.0 | 2.0 | 171.0 |
| 03 | 20.0 | 1.0 | 9.0 | 380.0 |
| 04 | 4.0 | 1.0 | 385.0 | 20.0 |
| 05 | 30.0 | 41.0 | 334.0 | 5.0 |
| 06 | 59.0 | 333.0 | 18.0 | 0.0 |
| 07 | 201.0 | 74.0 | 6.0 | 129.0 |
| 08 | 56.0 | 142.0 | 130.0 | 82.0 |
| 09 | 106.0 | 132.0 | 80.0 | 92.0 |
| 10 | 90.0 | 134.0 | 100.0 | 86.0 |
| 11 | 102.0 | 83.0 | 89.0 | 136.0 |
| 12 | 42.0 | 78.0 | 178.0 | 112.0 |
| 13 | 54.0 | 233.0 | 41.0 | 82.0 |
| 14 | 47.0 | 276.0 | 38.0 | 49.0 |
| 15 | 185.0 | 119.0 | 23.0 | 83.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.968 | 0.636 | -0.269 | -0.034 |
| 02 | -2.293 | 0.791 | -3.391 | 0.506 |
| 03 | -1.576 | -3.727 | -2.293 | 1.3 |
| 04 | -2.939 | -3.727 | 1.313 | -1.576 |
| 05 | -1.194 | -0.895 | 1.171 | -2.772 |
| 06 | -0.542 | 1.168 | -1.674 | -4.236 |
| 07 | 0.666 | -0.32 | -2.629 | 0.227 |
| 08 | -0.593 | 0.322 | 0.235 | -0.22 |
| 09 | 0.033 | 0.25 | -0.244 | -0.106 |
| 10 | -0.128 | 0.265 | -0.024 | -0.173 |
| 11 | -0.005 | -0.208 | -0.139 | 0.279 |
| 12 | -0.872 | -0.269 | 0.546 | 0.087 |
| 13 | -0.628 | 0.813 | -0.895 | -0.22 |
| 14 | -0.763 | 0.981 | -0.968 | -0.722 |
| 15 | 0.584 | 0.147 | -1.446 | -0.208 |