Model info
| Transcription factor | Nfyb | ||||||||
| Model | NFYB_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 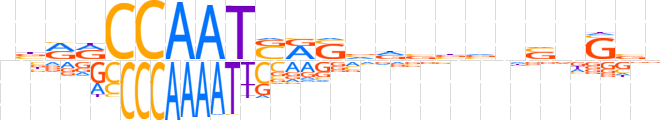 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bbRRCCAATSRSvdbbvnvnSbv | ||||||||
| Best auROC (human) | 0.987 | ||||||||
| Best auROC (mouse) | 0.998 | ||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | 4 | ||||||||
| Aligned words | 490 | ||||||||
| TF family | Heteromeric CCAAT-binding factors {4.2.1} | ||||||||
| TF subfamily | NF-YB (CP1B, CBF-A) {4.2.1.0.2} | ||||||||
| MGI | MGI:97317 | ||||||||
| EntrezGene | GeneID:18045 (SSTAR profile) | ||||||||
| UniProt ID | NFYB_MOUSE | ||||||||
| UniProt AC | P63139 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nfyb expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 7.069 | 22.302 | 26.01 | 4.186 | 9.889 | 72.724 | 20.747 | 56.028 | 8.815 | 67.05 | 21.469 | 17.969 | 5.246 | 50.58 | 72.608 | 26.641 |
| 02 | 13.885 | 0.0 | 17.135 | 0.0 | 122.574 | 20.378 | 61.153 | 8.551 | 44.062 | 9.096 | 87.677 | 0.0 | 32.755 | 6.174 | 64.976 | 0.918 |
| 03 | 39.163 | 5.736 | 152.736 | 15.641 | 21.45 | 1.209 | 6.349 | 6.64 | 84.667 | 8.028 | 116.307 | 21.938 | 5.368 | 0.0 | 1.728 | 2.373 |
| 04 | 0.999 | 148.808 | 0.841 | 0.0 | 0.0 | 12.98 | 1.14 | 0.853 | 0.0 | 277.12 | 0.0 | 0.0 | 0.0 | 44.389 | 2.204 | 0.0 |
| 05 | 0.0 | 0.999 | 0.0 | 0.0 | 0.713 | 481.831 | 0.753 | 0.0 | 0.0 | 4.185 | 0.0 | 0.0 | 0.0 | 0.853 | 0.0 | 0.0 |
| 06 | 0.0 | 0.713 | 0.0 | 0.0 | 483.497 | 0.0 | 0.0 | 4.371 | 0.753 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 474.554 | 3.699 | 4.886 | 1.111 | 0.713 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.371 | 0.0 | 0.0 | 0.0 |
| 08 | 0.729 | 13.323 | 0.0 | 465.586 | 0.0 | 0.0 | 0.0 | 3.699 | 0.0 | 0.0 | 0.0 | 4.886 | 0.0 | 0.0 | 0.0 | 1.111 |
| 09 | 0.0 | 0.0 | 0.729 | 0.0 | 0.945 | 11.52 | 0.858 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 35.671 | 289.378 | 143.238 | 6.995 |
| 10 | 14.111 | 0.0 | 22.505 | 0.0 | 218.029 | 28.039 | 53.788 | 1.041 | 52.716 | 1.245 | 87.706 | 3.159 | 2.813 | 0.0 | 4.182 | 0.0 |
| 11 | 28.223 | 35.768 | 214.87 | 8.807 | 3.54 | 13.047 | 10.684 | 2.013 | 22.641 | 28.6 | 114.278 | 2.664 | 0.0 | 1.041 | 3.159 | 0.0 |
| 12 | 30.062 | 7.788 | 15.359 | 1.195 | 17.986 | 24.755 | 19.667 | 16.048 | 146.191 | 116.244 | 62.49 | 18.064 | 3.069 | 2.327 | 6.129 | 1.959 |
| 13 | 51.734 | 11.686 | 96.555 | 37.334 | 64.871 | 2.152 | 59.357 | 24.735 | 46.399 | 12.057 | 30.874 | 14.315 | 3.995 | 2.044 | 27.263 | 3.963 |
| 14 | 18.769 | 22.699 | 111.012 | 14.519 | 5.613 | 2.9 | 13.984 | 5.441 | 29.951 | 87.131 | 68.609 | 28.359 | 4.575 | 15.124 | 37.024 | 23.624 |
| 15 | 14.236 | 17.047 | 21.054 | 6.57 | 8.722 | 40.252 | 44.178 | 34.701 | 13.363 | 102.602 | 82.559 | 32.107 | 6.514 | 21.194 | 26.692 | 17.544 |
| 16 | 8.176 | 7.877 | 25.551 | 1.231 | 34.028 | 65.429 | 55.518 | 26.119 | 54.106 | 74.266 | 38.826 | 7.285 | 14.179 | 42.399 | 29.281 | 5.063 |
| 17 | 28.843 | 9.396 | 35.7 | 36.551 | 34.118 | 31.825 | 67.778 | 56.252 | 38.472 | 42.302 | 52.719 | 15.682 | 3.785 | 7.503 | 20.197 | 8.212 |
| 18 | 8.519 | 14.537 | 79.504 | 2.659 | 4.854 | 37.262 | 36.932 | 11.977 | 30.553 | 61.615 | 74.794 | 9.432 | 3.577 | 29.784 | 73.395 | 9.942 |
| 19 | 7.873 | 1.974 | 30.082 | 7.574 | 36.43 | 31.977 | 38.173 | 36.617 | 69.999 | 47.858 | 81.015 | 65.753 | 3.602 | 4.681 | 15.986 | 9.741 |
| 20 | 7.133 | 6.352 | 103.503 | 0.916 | 13.632 | 9.848 | 54.473 | 8.539 | 13.655 | 22.025 | 117.62 | 11.955 | 0.0 | 11.256 | 104.617 | 3.811 |
| 21 | 8.315 | 3.727 | 20.474 | 1.904 | 2.719 | 18.552 | 9.684 | 18.527 | 48.801 | 103.459 | 185.34 | 42.613 | 2.241 | 6.045 | 14.883 | 2.051 |
| 22 | 15.788 | 10.287 | 28.418 | 7.583 | 27.141 | 28.162 | 46.257 | 30.223 | 53.144 | 49.56 | 110.627 | 17.049 | 6.808 | 18.853 | 28.991 | 10.442 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.424 | -0.311 | -0.16 | -1.913 | -1.103 | 0.859 | -0.382 | 0.6 | -1.214 | 0.778 | -0.349 | -0.523 | -1.704 | 0.498 | 0.857 | -0.136 |
| 02 | -0.775 | -4.382 | -0.57 | -4.382 | 1.379 | -0.4 | 0.687 | -1.243 | 0.361 | -1.184 | 1.045 | -4.382 | 0.068 | -1.552 | 0.747 | -3.167 |
| 03 | 0.245 | -1.621 | 1.598 | -0.659 | -0.349 | -2.966 | -1.526 | -1.483 | 1.01 | -1.303 | 1.327 | -0.327 | -1.683 | -4.382 | -2.684 | -2.418 |
| 04 | -3.106 | 1.572 | -3.228 | -4.382 | -4.382 | -0.84 | -3.01 | -3.218 | -4.382 | 2.193 | -4.382 | -4.382 | -4.382 | 0.369 | -2.481 | -4.382 |
| 05 | -4.382 | -3.106 | -4.382 | -4.382 | -3.337 | 2.745 | -3.302 | -4.382 | -4.382 | -1.913 | -4.382 | -4.382 | -4.382 | -3.218 | -4.382 | -4.382 |
| 06 | -4.382 | -3.337 | -4.382 | -4.382 | 2.749 | -4.382 | -4.382 | -1.873 | -3.302 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 |
| 07 | 2.73 | -2.025 | -1.77 | -3.029 | -3.337 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -1.873 | -4.382 | -4.382 | -4.382 |
| 08 | -3.323 | -0.815 | -4.382 | 2.711 | -4.382 | -4.382 | -4.382 | -2.025 | -4.382 | -4.382 | -4.382 | -1.77 | -4.382 | -4.382 | -4.382 | -3.029 |
| 09 | -4.382 | -4.382 | -3.323 | -4.382 | -3.146 | -0.956 | -3.213 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | 0.152 | 2.236 | 1.534 | -1.434 |
| 10 | -0.759 | -4.382 | -0.302 | -4.382 | 1.953 | -0.086 | 0.559 | -3.076 | 0.539 | -2.943 | 1.045 | -2.167 | -2.27 | -4.382 | -1.914 | -4.382 |
| 11 | -0.079 | 0.155 | 1.939 | -1.214 | -2.065 | -0.835 | -1.029 | -2.557 | -0.296 | -0.066 | 1.309 | -2.318 | -4.382 | -3.076 | -2.167 | -4.382 |
| 12 | -0.017 | -1.332 | -0.676 | -2.974 | -0.522 | -0.208 | -0.435 | -0.634 | 1.555 | 1.326 | 0.708 | -0.518 | -2.193 | -2.435 | -1.559 | -2.58 |
| 13 | 0.521 | -0.942 | 1.141 | 0.197 | 0.745 | -2.501 | 0.657 | -0.209 | 0.413 | -0.912 | 0.009 | -0.745 | -1.955 | -2.545 | -0.113 | -1.963 |
| 14 | -0.48 | -0.294 | 1.28 | -0.731 | -1.641 | -2.243 | -0.768 | -1.67 | -0.021 | 1.039 | 0.801 | -0.075 | -1.831 | -0.691 | 0.189 | -0.255 |
| 15 | -0.75 | -0.575 | -0.368 | -1.493 | -1.224 | 0.272 | 0.364 | 0.125 | -0.812 | 1.202 | 0.985 | 0.048 | -1.501 | -0.361 | -0.134 | -0.546 |
| 16 | -1.286 | -1.321 | -0.177 | -2.952 | 0.105 | 0.754 | 0.591 | -0.156 | 0.565 | 0.88 | 0.236 | -1.395 | -0.754 | 0.323 | -0.043 | -1.737 |
| 17 | -0.058 | -1.152 | 0.153 | 0.176 | 0.108 | 0.039 | 0.789 | 0.604 | 0.227 | 0.321 | 0.539 | -0.656 | -2.005 | -1.367 | -0.409 | -1.281 |
| 18 | -1.246 | -0.73 | 0.948 | -2.319 | -1.776 | 0.195 | 0.186 | -0.918 | -0.001 | 0.694 | 0.887 | -1.149 | -2.056 | -0.026 | 0.868 | -1.098 |
| 19 | -1.322 | -2.574 | -0.016 | -1.359 | 0.173 | 0.044 | 0.219 | 0.178 | 0.821 | 0.443 | 0.966 | 0.759 | -2.05 | -1.81 | -0.637 | -1.118 |
| 20 | -1.415 | -1.525 | 1.21 | -3.168 | -0.793 | -1.107 | 0.572 | -1.244 | -0.791 | -0.323 | 1.338 | -0.92 | -4.382 | -0.978 | 1.221 | -1.998 |
| 21 | -1.27 | -2.019 | -0.395 | -2.604 | -2.3 | -0.492 | -1.123 | -0.493 | 0.463 | 1.21 | 1.791 | 0.328 | -2.467 | -1.572 | -0.707 | -2.542 |
| 22 | -0.65 | -1.065 | -0.072 | -1.357 | -0.118 | -0.081 | 0.41 | -0.012 | 0.547 | 0.478 | 1.277 | -0.574 | -1.46 | -0.476 | -0.053 | -1.051 |