Model info
| Transcription factor | NKX2-5 (GeneCards) | ||||||||
| Model | NKX25_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 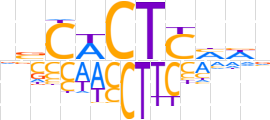 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 10 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nhdGAGWGbn | ||||||||
| Best auROC (human) | 0.749 | ||||||||
| Best auROC (mouse) | 0.889 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 7 | ||||||||
| Aligned words | 518 | ||||||||
| TF family | NK-related factors {3.1.2} | ||||||||
| TF subfamily | NK-4 {3.1.2.17} | ||||||||
| HGNC | HGNC:2488 | ||||||||
| EntrezGene | GeneID:1482 (SSTAR profile) | ||||||||
| UniProt ID | NKX25_HUMAN | ||||||||
| UniProt AC | P52952 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | NKX2-5 expression | ||||||||
| ReMap ChIP-seq dataset list | NKX2-5 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 9.0 | 9.0 | 16.0 | 37.0 | 40.0 | 12.0 | 4.0 | 116.0 | 37.0 | 25.0 | 12.0 | 63.0 | 14.0 | 12.0 | 17.0 | 71.0 |
| 02 | 10.0 | 3.0 | 34.0 | 53.0 | 16.0 | 6.0 | 1.0 | 35.0 | 7.0 | 3.0 | 10.0 | 29.0 | 27.0 | 15.0 | 95.0 | 150.0 |
| 03 | 6.0 | 3.0 | 51.0 | 0.0 | 18.0 | 2.0 | 5.0 | 2.0 | 38.0 | 1.0 | 101.0 | 0.0 | 43.0 | 3.0 | 221.0 | 0.0 |
| 04 | 104.0 | 0.0 | 0.0 | 1.0 | 9.0 | 0.0 | 0.0 | 0.0 | 375.0 | 2.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 05 | 2.0 | 0.0 | 487.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 165.0 | 2.0 | 16.0 | 308.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 07 | 8.0 | 0.0 | 148.0 | 9.0 | 0.0 | 0.0 | 0.0 | 2.0 | 2.0 | 1.0 | 13.0 | 0.0 | 28.0 | 1.0 | 276.0 | 6.0 |
| 08 | 0.0 | 22.0 | 13.0 | 3.0 | 0.0 | 2.0 | 0.0 | 0.0 | 46.0 | 121.0 | 214.0 | 56.0 | 2.0 | 6.0 | 9.0 | 0.0 |
| 09 | 6.0 | 15.0 | 13.0 | 14.0 | 30.0 | 40.0 | 6.0 | 75.0 | 51.0 | 94.0 | 66.0 | 25.0 | 16.0 | 12.0 | 14.0 | 17.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.203 | -1.203 | -0.646 | 0.179 | 0.256 | -0.926 | -1.964 | 1.315 | 0.179 | -0.208 | -0.926 | 0.707 | -0.776 | -0.926 | -0.587 | 0.826 |
| 02 | -1.102 | -2.222 | 0.095 | 0.535 | -0.646 | -1.588 | -3.115 | 0.124 | -1.443 | -2.222 | -1.102 | -0.062 | -0.132 | -0.709 | 1.116 | 1.571 |
| 03 | -1.588 | -2.222 | 0.497 | -4.39 | -0.531 | -2.572 | -1.758 | -2.572 | 0.205 | -3.115 | 1.177 | -4.39 | 0.328 | -2.222 | 1.957 | -4.39 |
| 04 | 1.206 | -4.39 | -4.39 | -3.115 | -1.203 | -4.39 | -4.39 | -4.39 | 2.486 | -2.572 | -4.39 | -3.115 | -2.572 | -4.39 | -4.39 | -4.39 |
| 05 | -2.572 | -4.39 | 2.747 | -3.115 | -4.39 | -4.39 | -2.572 | -4.39 | -4.39 | -4.39 | -4.39 | -4.39 | -4.39 | -4.39 | -2.572 | -4.39 |
| 06 | -4.39 | -4.39 | -4.39 | -2.572 | -4.39 | -4.39 | -4.39 | -4.39 | 1.666 | -2.572 | -0.646 | 2.289 | -4.39 | -4.39 | -4.39 | -3.115 |
| 07 | -1.316 | -4.39 | 1.557 | -1.203 | -4.39 | -4.39 | -4.39 | -2.572 | -2.572 | -3.115 | -0.848 | -4.39 | -0.096 | -3.115 | 2.179 | -1.588 |
| 08 | -4.39 | -0.334 | -0.848 | -2.222 | -4.39 | -2.572 | -4.39 | -4.39 | 0.395 | 1.357 | 1.925 | 0.59 | -2.572 | -1.588 | -1.203 | -4.39 |
| 09 | -1.588 | -0.709 | -0.848 | -0.776 | -0.028 | 0.256 | -1.588 | 0.88 | 0.497 | 1.105 | 0.753 | -0.208 | -0.646 | -0.926 | -0.776 | -0.587 |