Model info
| Transcription factor | Nkx3-2 | ||||||||
| Model | NKX32_MOUSE.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 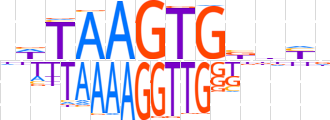 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nWTAAGTGbdYn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.979 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 6 | ||||||||
| Aligned words | 585 | ||||||||
| TF family | NK-related factors {3.1.2} | ||||||||
| TF subfamily | NK-3 {3.1.2.16} | ||||||||
| MGI | MGI:108015 | ||||||||
| EntrezGene | GeneID:12020 (SSTAR profile) | ||||||||
| UniProt ID | NKX32_MOUSE | ||||||||
| UniProt AC | P97503 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nkx3-2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 22.0 | 22.0 | 10.0 | 82.0 | 16.0 | 11.0 | 4.0 | 48.0 | 27.0 | 12.0 | 5.0 | 58.0 | 36.0 | 7.0 | 27.0 | 98.0 |
| 02 | 0.0 | 6.0 | 2.0 | 93.0 | 0.0 | 4.0 | 0.0 | 48.0 | 2.0 | 2.0 | 4.0 | 38.0 | 15.0 | 7.0 | 9.0 | 255.0 |
| 03 | 17.0 | 0.0 | 0.0 | 0.0 | 18.0 | 1.0 | 0.0 | 0.0 | 14.0 | 1.0 | 0.0 | 0.0 | 411.0 | 11.0 | 12.0 | 0.0 |
| 04 | 458.0 | 1.0 | 0.0 | 1.0 | 12.0 | 1.0 | 0.0 | 0.0 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 481.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 15.0 | 467.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 07 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 13.0 | 0.0 | 9.0 | 2.0 | 457.0 | 0.0 |
| 08 | 0.0 | 4.0 | 3.0 | 3.0 | 0.0 | 2.0 | 1.0 | 0.0 | 19.0 | 83.0 | 157.0 | 213.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 4.0 | 3.0 | 6.0 | 6.0 | 23.0 | 11.0 | 5.0 | 50.0 | 58.0 | 22.0 | 18.0 | 63.0 | 43.0 | 26.0 | 72.0 | 75.0 |
| 10 | 19.0 | 32.0 | 8.0 | 69.0 | 14.0 | 16.0 | 2.0 | 30.0 | 10.0 | 13.0 | 4.0 | 74.0 | 33.0 | 31.0 | 15.0 | 115.0 |
| 11 | 19.0 | 20.0 | 20.0 | 17.0 | 37.0 | 10.0 | 5.0 | 40.0 | 7.0 | 5.0 | 8.0 | 9.0 | 79.0 | 33.0 | 102.0 | 74.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.316 | -0.316 | -1.084 | 0.987 | -0.628 | -0.992 | -1.946 | 0.455 | -0.114 | -0.908 | -1.74 | 0.643 | 0.17 | -1.425 | -0.114 | 1.165 |
| 02 | -4.375 | -1.57 | -2.554 | 1.113 | -4.375 | -1.946 | -4.375 | 0.455 | -2.554 | -2.554 | -1.946 | 0.223 | -0.691 | -1.425 | -1.185 | 2.119 |
| 03 | -0.569 | -4.375 | -4.375 | -4.375 | -0.513 | -3.097 | -4.375 | -4.375 | -0.758 | -3.097 | -4.375 | -4.375 | 2.595 | -0.992 | -0.908 | -4.375 |
| 04 | 2.703 | -3.097 | -4.375 | -3.097 | -0.908 | -3.097 | -4.375 | -4.375 | -0.908 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 |
| 05 | -4.375 | -4.375 | 2.752 | -3.097 | -4.375 | -4.375 | -2.554 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -3.097 | -4.375 |
| 06 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -2.554 | -4.375 | -0.691 | 2.723 | -4.375 | -4.375 | -4.375 | -3.097 |
| 07 | -4.375 | -4.375 | -2.554 | -4.375 | -4.375 | -4.375 | -4.375 | -4.375 | -3.097 | -3.097 | -0.83 | -4.375 | -1.185 | -2.554 | 2.701 | -4.375 |
| 08 | -4.375 | -1.946 | -2.204 | -2.204 | -4.375 | -2.554 | -3.097 | -4.375 | -0.46 | 0.999 | 1.634 | 1.939 | -4.375 | -4.375 | -4.375 | -4.375 |
| 09 | -1.946 | -2.204 | -1.57 | -1.57 | -0.272 | -0.992 | -1.74 | 0.495 | 0.643 | -0.316 | -0.513 | 0.725 | 0.346 | -0.151 | 0.858 | 0.898 |
| 10 | -0.46 | 0.054 | -1.298 | 0.815 | -0.758 | -0.628 | -2.554 | -0.01 | -1.084 | -0.83 | -1.946 | 0.885 | 0.084 | 0.022 | -0.691 | 1.324 |
| 11 | -0.46 | -0.409 | -0.409 | -0.569 | 0.197 | -1.084 | -1.74 | 0.274 | -1.425 | -1.74 | -1.298 | -1.185 | 0.95 | 0.084 | 1.205 | 0.885 |