Model info
| Transcription factor | Nr1h4 | ||||||||
| Model | NR1H4_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 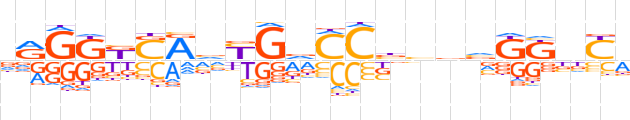 | ||||||||
| LOGO (reverse complement) | 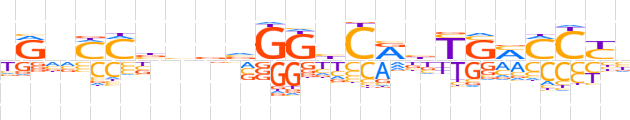 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vRGKKSRvYGdCCbvbvKKbYv | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.942 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 15 | ||||||||
| Aligned words | 472 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | LXR (NR1H) {2.1.2.7} | ||||||||
| MGI | MGI:1352464 | ||||||||
| EntrezGene | GeneID:20186 (SSTAR profile) | ||||||||
| UniProt ID | NR1H4_MOUSE | ||||||||
| UniProt AC | Q60641 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nr1h4 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 53.0 | 2.0 | 68.0 | 7.0 | 46.0 | 2.0 | 36.0 | 5.0 | 86.0 | 3.0 | 99.0 | 7.0 | 13.0 | 4.0 | 41.0 | 0.0 |
| 02 | 11.0 | 2.0 | 176.0 | 9.0 | 4.0 | 0.0 | 7.0 | 0.0 | 31.0 | 6.0 | 194.0 | 13.0 | 3.0 | 1.0 | 14.0 | 1.0 |
| 03 | 2.0 | 3.0 | 43.0 | 1.0 | 2.0 | 0.0 | 7.0 | 0.0 | 34.0 | 10.0 | 290.0 | 57.0 | 0.0 | 1.0 | 20.0 | 2.0 |
| 04 | 3.0 | 1.0 | 26.0 | 8.0 | 0.0 | 0.0 | 2.0 | 12.0 | 21.0 | 39.0 | 84.0 | 216.0 | 0.0 | 3.0 | 19.0 | 38.0 |
| 05 | 0.0 | 20.0 | 3.0 | 1.0 | 0.0 | 36.0 | 2.0 | 5.0 | 3.0 | 104.0 | 8.0 | 16.0 | 2.0 | 188.0 | 54.0 | 30.0 |
| 06 | 2.0 | 0.0 | 3.0 | 0.0 | 283.0 | 8.0 | 49.0 | 8.0 | 52.0 | 8.0 | 5.0 | 2.0 | 21.0 | 13.0 | 17.0 | 1.0 |
| 07 | 157.0 | 89.0 | 82.0 | 30.0 | 11.0 | 5.0 | 8.0 | 5.0 | 23.0 | 26.0 | 16.0 | 9.0 | 2.0 | 4.0 | 5.0 | 0.0 |
| 08 | 14.0 | 45.0 | 27.0 | 107.0 | 3.0 | 17.0 | 4.0 | 100.0 | 8.0 | 25.0 | 10.0 | 68.0 | 0.0 | 7.0 | 1.0 | 36.0 |
| 09 | 1.0 | 3.0 | 21.0 | 0.0 | 20.0 | 1.0 | 72.0 | 1.0 | 9.0 | 3.0 | 29.0 | 1.0 | 21.0 | 7.0 | 281.0 | 2.0 |
| 10 | 18.0 | 5.0 | 26.0 | 2.0 | 10.0 | 0.0 | 1.0 | 3.0 | 187.0 | 54.0 | 106.0 | 56.0 | 2.0 | 1.0 | 1.0 | 0.0 |
| 11 | 26.0 | 182.0 | 6.0 | 3.0 | 6.0 | 44.0 | 3.0 | 7.0 | 6.0 | 109.0 | 8.0 | 11.0 | 2.0 | 49.0 | 3.0 | 7.0 |
| 12 | 1.0 | 36.0 | 2.0 | 1.0 | 30.0 | 328.0 | 1.0 | 25.0 | 1.0 | 17.0 | 1.0 | 1.0 | 4.0 | 23.0 | 1.0 | 0.0 |
| 13 | 0.0 | 20.0 | 12.0 | 4.0 | 19.0 | 123.0 | 126.0 | 136.0 | 0.0 | 1.0 | 2.0 | 2.0 | 0.0 | 14.0 | 10.0 | 3.0 |
| 14 | 1.0 | 5.0 | 11.0 | 2.0 | 42.0 | 57.0 | 22.0 | 37.0 | 19.0 | 84.0 | 35.0 | 12.0 | 18.0 | 55.0 | 50.0 | 22.0 |
| 15 | 11.0 | 13.0 | 51.0 | 5.0 | 46.0 | 48.0 | 57.0 | 50.0 | 17.0 | 35.0 | 47.0 | 19.0 | 9.0 | 14.0 | 39.0 | 11.0 |
| 16 | 20.0 | 20.0 | 42.0 | 1.0 | 49.0 | 29.0 | 20.0 | 12.0 | 54.0 | 72.0 | 57.0 | 11.0 | 8.0 | 18.0 | 53.0 | 6.0 |
| 17 | 10.0 | 4.0 | 114.0 | 3.0 | 12.0 | 1.0 | 109.0 | 17.0 | 22.0 | 5.0 | 114.0 | 31.0 | 2.0 | 1.0 | 22.0 | 5.0 |
| 18 | 6.0 | 4.0 | 35.0 | 1.0 | 4.0 | 1.0 | 3.0 | 3.0 | 25.0 | 31.0 | 272.0 | 31.0 | 4.0 | 3.0 | 41.0 | 8.0 |
| 19 | 5.0 | 11.0 | 19.0 | 4.0 | 15.0 | 7.0 | 9.0 | 8.0 | 33.0 | 93.0 | 95.0 | 130.0 | 2.0 | 12.0 | 8.0 | 21.0 |
| 20 | 0.0 | 44.0 | 6.0 | 5.0 | 3.0 | 82.0 | 12.0 | 26.0 | 3.0 | 103.0 | 12.0 | 13.0 | 2.0 | 120.0 | 15.0 | 26.0 |
| 21 | 1.0 | 5.0 | 0.0 | 2.0 | 226.0 | 28.0 | 60.0 | 35.0 | 12.0 | 11.0 | 14.0 | 8.0 | 21.0 | 12.0 | 30.0 | 7.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.58 | -2.528 | 0.828 | -1.398 | 0.44 | -2.528 | 0.197 | -1.714 | 1.061 | -2.178 | 1.202 | -1.398 | -0.803 | -1.919 | 0.326 | -4.352 |
| 02 | -0.965 | -2.528 | 1.775 | -1.158 | -1.919 | -4.352 | -1.398 | -4.352 | 0.049 | -1.543 | 1.872 | -0.803 | -2.178 | -3.072 | -0.731 | -3.072 |
| 03 | -2.528 | -2.178 | 0.373 | -3.072 | -2.528 | -4.352 | -1.398 | -4.352 | 0.14 | -1.057 | 2.274 | 0.652 | -4.352 | -3.072 | -0.383 | -2.528 |
| 04 | -2.178 | -3.072 | -0.125 | -1.271 | -4.352 | -4.352 | -2.528 | -0.881 | -0.335 | 0.276 | 1.038 | 1.98 | -4.352 | -2.178 | -0.433 | 0.25 |
| 05 | -4.352 | -0.383 | -2.178 | -3.072 | -4.352 | 0.197 | -2.528 | -1.714 | -2.178 | 1.251 | -1.271 | -0.601 | -2.528 | 1.841 | 0.599 | 0.017 |
| 06 | -2.528 | -4.352 | -2.178 | -4.352 | 2.249 | -1.271 | 0.502 | -1.271 | 0.561 | -1.271 | -1.714 | -2.528 | -0.335 | -0.803 | -0.542 | -3.072 |
| 07 | 1.661 | 1.096 | 1.014 | 0.017 | -0.965 | -1.714 | -1.271 | -1.714 | -0.245 | -0.125 | -0.601 | -1.158 | -2.528 | -1.919 | -1.714 | -4.352 |
| 08 | -0.731 | 0.418 | -0.087 | 1.279 | -2.178 | -0.542 | -1.919 | 1.212 | -1.271 | -0.163 | -1.057 | 0.828 | -4.352 | -1.398 | -3.072 | 0.197 |
| 09 | -3.072 | -2.178 | -0.335 | -4.352 | -0.383 | -3.072 | 0.885 | -3.072 | -1.158 | -2.178 | -0.017 | -3.072 | -0.335 | -1.398 | 2.242 | -2.528 |
| 10 | -0.486 | -1.714 | -0.125 | -2.528 | -1.057 | -4.352 | -3.072 | -2.178 | 1.836 | 0.599 | 1.27 | 0.635 | -2.528 | -3.072 | -3.072 | -4.352 |
| 11 | -0.125 | 1.809 | -1.543 | -2.178 | -1.543 | 0.396 | -2.178 | -1.398 | -1.543 | 1.298 | -1.271 | -0.965 | -2.528 | 0.502 | -2.178 | -1.398 |
| 12 | -3.072 | 0.197 | -2.528 | -3.072 | 0.017 | 2.397 | -3.072 | -0.163 | -3.072 | -0.542 | -3.072 | -3.072 | -1.919 | -0.245 | -3.072 | -4.352 |
| 13 | -4.352 | -0.383 | -0.881 | -1.919 | -0.433 | 1.418 | 1.442 | 1.518 | -4.352 | -3.072 | -2.528 | -2.528 | -4.352 | -0.731 | -1.057 | -2.178 |
| 14 | -3.072 | -1.714 | -0.965 | -2.528 | 0.349 | 0.652 | -0.289 | 0.224 | -0.433 | 1.038 | 0.169 | -0.881 | -0.486 | 0.617 | 0.522 | -0.289 |
| 15 | -0.965 | -0.803 | 0.542 | -1.714 | 0.44 | 0.482 | 0.652 | 0.522 | -0.542 | 0.169 | 0.461 | -0.433 | -1.158 | -0.731 | 0.276 | -0.965 |
| 16 | -0.383 | -0.383 | 0.349 | -3.072 | 0.502 | -0.017 | -0.383 | -0.881 | 0.599 | 0.885 | 0.652 | -0.965 | -1.271 | -0.486 | 0.58 | -1.543 |
| 17 | -1.057 | -1.919 | 1.342 | -2.178 | -0.881 | -3.072 | 1.298 | -0.542 | -0.289 | -1.714 | 1.342 | 0.049 | -2.528 | -3.072 | -0.289 | -1.714 |
| 18 | -1.543 | -1.919 | 0.169 | -3.072 | -1.919 | -3.072 | -2.178 | -2.178 | -0.163 | 0.049 | 2.21 | 0.049 | -1.919 | -2.178 | 0.326 | -1.271 |
| 19 | -1.714 | -0.965 | -0.433 | -1.919 | -0.664 | -1.398 | -1.158 | -1.271 | 0.111 | 1.139 | 1.161 | 1.473 | -2.528 | -0.881 | -1.271 | -0.335 |
| 20 | -4.352 | 0.396 | -1.543 | -1.714 | -2.178 | 1.014 | -0.881 | -0.125 | -2.178 | 1.241 | -0.881 | -0.803 | -2.528 | 1.393 | -0.664 | -0.125 |
| 21 | -3.072 | -1.714 | -4.352 | -2.528 | 2.025 | -0.051 | 0.703 | 0.169 | -0.881 | -0.965 | -0.731 | -1.271 | -0.335 | -0.881 | 0.017 | -1.398 |