Model info
| Transcription factor | NR2C2 (GeneCards) | ||||||||
| Model | NR2C2_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 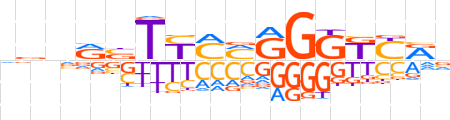 | ||||||||
| LOGO (reverse complement) | 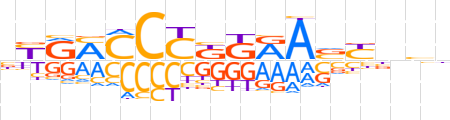 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dbnRbTYMMGGGKYRv | ||||||||
| Best auROC (human) | 0.885 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 35 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 556 | ||||||||
| TF family | RXR-related receptors (NR2) {2.1.3} | ||||||||
| TF subfamily | Testicular receptors (NR2C) {2.1.3.4} | ||||||||
| HGNC | HGNC:7972 | ||||||||
| EntrezGene | GeneID:7182 (SSTAR profile) | ||||||||
| UniProt ID | NR2C2_HUMAN | ||||||||
| UniProt AC | P49116 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | NR2C2 expression | ||||||||
| ReMap ChIP-seq dataset list | NR2C2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 20.0 | 37.0 | 116.0 | 20.0 | 12.0 | 21.0 | 25.0 | 7.0 | 21.0 | 9.0 | 38.0 | 18.0 | 21.0 | 40.0 | 41.0 | 48.0 |
| 02 | 23.0 | 9.0 | 35.0 | 7.0 | 31.0 | 45.0 | 8.0 | 23.0 | 73.0 | 48.0 | 63.0 | 36.0 | 12.0 | 49.0 | 7.0 | 25.0 |
| 03 | 23.0 | 7.0 | 107.0 | 2.0 | 71.0 | 19.0 | 49.0 | 12.0 | 14.0 | 2.0 | 94.0 | 3.0 | 35.0 | 9.0 | 41.0 | 6.0 |
| 04 | 3.0 | 97.0 | 24.0 | 19.0 | 10.0 | 17.0 | 4.0 | 6.0 | 7.0 | 72.0 | 176.0 | 36.0 | 1.0 | 15.0 | 5.0 | 2.0 |
| 05 | 0.0 | 0.0 | 2.0 | 19.0 | 13.0 | 4.0 | 4.0 | 180.0 | 3.0 | 5.0 | 10.0 | 191.0 | 2.0 | 0.0 | 2.0 | 59.0 |
| 06 | 0.0 | 6.0 | 0.0 | 12.0 | 0.0 | 1.0 | 0.0 | 8.0 | 1.0 | 10.0 | 1.0 | 6.0 | 2.0 | 137.0 | 23.0 | 287.0 |
| 07 | 2.0 | 1.0 | 0.0 | 0.0 | 129.0 | 13.0 | 11.0 | 1.0 | 18.0 | 6.0 | 0.0 | 0.0 | 28.0 | 281.0 | 3.0 | 1.0 |
| 08 | 99.0 | 4.0 | 73.0 | 1.0 | 6.0 | 282.0 | 12.0 | 1.0 | 5.0 | 1.0 | 8.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 09 | 75.0 | 0.0 | 36.0 | 1.0 | 31.0 | 0.0 | 256.0 | 0.0 | 58.0 | 0.0 | 34.0 | 1.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 10 | 2.0 | 1.0 | 156.0 | 7.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 6.0 | 312.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 11 | 0.0 | 0.0 | 9.0 | 0.0 | 1.0 | 0.0 | 5.0 | 1.0 | 18.0 | 11.0 | 371.0 | 70.0 | 0.0 | 0.0 | 2.0 | 6.0 |
| 12 | 2.0 | 2.0 | 6.0 | 9.0 | 1.0 | 0.0 | 6.0 | 4.0 | 7.0 | 10.0 | 99.0 | 271.0 | 1.0 | 9.0 | 17.0 | 50.0 |
| 13 | 1.0 | 4.0 | 5.0 | 1.0 | 1.0 | 16.0 | 1.0 | 3.0 | 4.0 | 78.0 | 24.0 | 22.0 | 4.0 | 262.0 | 28.0 | 40.0 |
| 14 | 5.0 | 2.0 | 1.0 | 2.0 | 236.0 | 21.0 | 99.0 | 4.0 | 34.0 | 17.0 | 7.0 | 0.0 | 28.0 | 21.0 | 12.0 | 5.0 |
| 15 | 69.0 | 74.0 | 120.0 | 40.0 | 7.0 | 13.0 | 26.0 | 15.0 | 11.0 | 41.0 | 54.0 | 13.0 | 6.0 | 1.0 | 1.0 | 3.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.427 | 0.179 | 1.315 | -0.427 | -0.926 | -0.38 | -0.208 | -1.443 | -0.38 | -1.203 | 0.205 | -0.531 | -0.38 | 0.256 | 0.281 | 0.437 |
| 02 | -0.29 | -1.203 | 0.124 | -1.443 | 0.004 | 0.373 | -1.316 | -0.29 | 0.853 | 0.437 | 0.707 | 0.152 | -0.926 | 0.457 | -1.443 | -0.208 |
| 03 | -0.29 | -1.443 | 1.234 | -2.572 | 0.826 | -0.478 | 0.457 | -0.926 | -0.776 | -2.572 | 1.105 | -2.222 | 0.124 | -1.203 | 0.281 | -1.588 |
| 04 | -2.222 | 1.136 | -0.248 | -0.478 | -1.102 | -0.587 | -1.964 | -1.588 | -1.443 | 0.84 | 1.73 | 0.152 | -3.115 | -0.709 | -1.758 | -2.572 |
| 05 | -4.39 | -4.39 | -2.572 | -0.478 | -0.848 | -1.964 | -1.964 | 1.753 | -2.222 | -1.758 | -1.102 | 1.812 | -2.572 | -4.39 | -2.572 | 0.642 |
| 06 | -4.39 | -1.588 | -4.39 | -0.926 | -4.39 | -3.115 | -4.39 | -1.316 | -3.115 | -1.102 | -3.115 | -1.588 | -2.572 | 1.48 | -0.29 | 2.218 |
| 07 | -2.572 | -3.115 | -4.39 | -4.39 | 1.42 | -0.848 | -1.01 | -3.115 | -0.531 | -1.588 | -4.39 | -4.39 | -0.096 | 2.197 | -2.222 | -3.115 |
| 08 | 1.157 | -1.964 | 0.853 | -3.115 | -1.588 | 2.201 | -0.926 | -3.115 | -1.758 | -3.115 | -1.316 | -4.39 | -2.572 | -4.39 | -4.39 | -4.39 |
| 09 | 0.88 | -4.39 | 0.152 | -3.115 | 0.004 | -4.39 | 2.104 | -4.39 | 0.625 | -4.39 | 0.095 | -3.115 | -2.572 | -4.39 | -4.39 | -4.39 |
| 10 | -2.572 | -3.115 | 1.61 | -1.443 | -4.39 | -4.39 | -4.39 | -4.39 | -1.443 | -1.588 | 2.302 | -3.115 | -4.39 | -4.39 | -2.572 | -4.39 |
| 11 | -4.39 | -4.39 | -1.203 | -4.39 | -3.115 | -4.39 | -1.758 | -3.115 | -0.531 | -1.01 | 2.475 | 0.812 | -4.39 | -4.39 | -2.572 | -1.588 |
| 12 | -2.572 | -2.572 | -1.588 | -1.203 | -3.115 | -4.39 | -1.588 | -1.964 | -1.443 | -1.102 | 1.157 | 2.161 | -3.115 | -1.203 | -0.587 | 0.477 |
| 13 | -3.115 | -1.964 | -1.758 | -3.115 | -3.115 | -0.646 | -3.115 | -2.222 | -1.964 | 0.919 | -0.248 | -0.334 | -1.964 | 2.127 | -0.096 | 0.256 |
| 14 | -1.758 | -2.572 | -3.115 | -2.572 | 2.023 | -0.38 | 1.157 | -1.964 | 0.095 | -0.587 | -1.443 | -4.39 | -0.096 | -0.38 | -0.926 | -1.758 |
| 15 | 0.797 | 0.867 | 1.348 | 0.256 | -1.443 | -0.848 | -0.17 | -0.709 | -1.01 | 0.281 | 0.554 | -0.848 | -1.588 | -3.115 | -3.115 | -2.222 |