Model info
| Transcription factor | Nr4a1 | ||||||||
| Model | NR4A1_MOUSE.H11MO.0.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 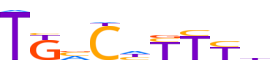 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 9 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dRARdGbMA | ||||||||
| Best auROC (human) | 0.903 | ||||||||
| Best auROC (mouse) | 0.631 | ||||||||
| Peak sets in benchmark (human) | 12 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 499 | ||||||||
| TF family | NGFI-B-related receptors (NR4) {2.1.4} | ||||||||
| TF subfamily | NGFI-B (Nur77, NR4A1) {2.1.4.0.1} | ||||||||
| MGI | MGI:1352454 | ||||||||
| EntrezGene | GeneID:15370 (SSTAR profile) | ||||||||
| UniProt ID | NR4A1_MOUSE | ||||||||
| UniProt AC | P12813 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nr4a1 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 206.0 | 74.0 | 134.0 | 85.0 |
| 02 | 311.0 | 22.0 | 146.0 | 20.0 |
| 03 | 379.0 | 12.0 | 103.0 | 5.0 |
| 04 | 361.0 | 59.0 | 72.0 | 7.0 |
| 05 | 169.0 | 11.0 | 184.0 | 135.0 |
| 06 | 37.0 | 7.0 | 436.0 | 19.0 |
| 07 | 26.0 | 69.0 | 190.0 | 214.0 |
| 08 | 92.0 | 368.0 | 33.0 | 6.0 |
| 09 | 475.0 | 2.0 | 4.0 | 18.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.497 | -0.514 | 0.071 | -0.378 |
| 02 | 0.906 | -1.679 | 0.156 | -1.768 |
| 03 | 1.103 | -2.232 | -0.189 | -2.959 |
| 04 | 1.054 | -0.735 | -0.541 | -2.692 |
| 05 | 0.3 | -2.309 | 0.385 | 0.078 |
| 06 | -1.187 | -2.692 | 1.243 | -1.816 |
| 07 | -1.523 | -0.582 | 0.416 | 0.535 |
| 08 | -0.3 | 1.074 | -1.296 | -2.817 |
| 09 | 1.328 | -3.571 | -3.124 | -1.866 |