Model info
| Transcription factor | Nrf1 | ||||||||
| Model | NRF1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 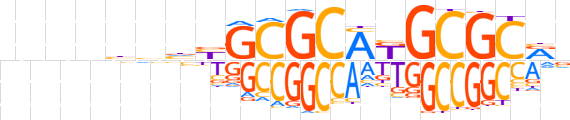 | ||||||||
| LOGO (reverse complement) | 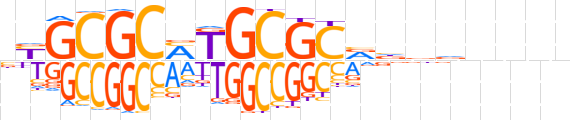 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnnnbvnbGCGCAKGCGCRv | ||||||||
| Best auROC (human) | 0.999 | ||||||||
| Best auROC (mouse) | 0.999 | ||||||||
| Peak sets in benchmark (human) | 31 | ||||||||
| Peak sets in benchmark (mouse) | 23 | ||||||||
| Aligned words | 407 | ||||||||
| TF family | NRF {0.0.6} | ||||||||
| TF subfamily | NRF-1 (alpha-pal) {0.0.6.0.1} | ||||||||
| MGI | MGI:1332235 | ||||||||
| EntrezGene | GeneID:18181 (SSTAR profile) | ||||||||
| UniProt ID | NRF1_MOUSE | ||||||||
| UniProt AC | Q9WU00 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nrf1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 30.0 | 32.0 | 27.0 | 12.0 | 21.0 | 45.0 | 12.0 | 23.0 | 31.0 | 48.0 | 18.0 | 26.0 | 16.0 | 26.0 | 15.0 | 24.0 |
| 02 | 16.0 | 25.0 | 47.0 | 10.0 | 37.0 | 48.0 | 35.0 | 31.0 | 22.0 | 17.0 | 18.0 | 15.0 | 12.0 | 31.0 | 32.0 | 10.0 |
| 03 | 23.0 | 27.0 | 27.0 | 10.0 | 13.0 | 36.0 | 31.0 | 41.0 | 27.0 | 44.0 | 43.0 | 18.0 | 7.0 | 25.0 | 23.0 | 11.0 |
| 04 | 12.0 | 24.0 | 21.0 | 13.0 | 18.0 | 55.0 | 14.0 | 45.0 | 25.0 | 64.0 | 19.0 | 16.0 | 3.0 | 44.0 | 18.0 | 15.0 |
| 05 | 4.0 | 29.0 | 22.0 | 3.0 | 66.0 | 47.0 | 17.0 | 57.0 | 3.0 | 39.0 | 18.0 | 12.0 | 12.0 | 51.0 | 22.0 | 4.0 |
| 06 | 17.0 | 40.0 | 24.0 | 4.0 | 42.0 | 74.0 | 10.0 | 40.0 | 14.0 | 34.0 | 18.0 | 13.0 | 4.0 | 57.0 | 10.0 | 5.0 |
| 07 | 9.0 | 26.0 | 15.0 | 27.0 | 6.0 | 51.0 | 15.0 | 133.0 | 7.0 | 9.0 | 8.0 | 38.0 | 3.0 | 17.0 | 16.0 | 26.0 |
| 08 | 1.0 | 0.0 | 23.0 | 1.0 | 26.0 | 3.0 | 74.0 | 0.0 | 9.0 | 2.0 | 43.0 | 0.0 | 20.0 | 1.0 | 203.0 | 0.0 |
| 09 | 3.0 | 51.0 | 2.0 | 0.0 | 0.0 | 6.0 | 0.0 | 0.0 | 38.0 | 300.0 | 5.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 41.0 | 0.0 | 15.0 | 2.0 | 340.0 | 1.0 | 2.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 0.0 | 15.0 | 2.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 377.0 | 3.0 | 5.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 12 | 0.0 | 0.0 | 0.0 | 1.0 | 330.0 | 41.0 | 6.0 | 17.0 | 1.0 | 3.0 | 0.0 | 1.0 | 0.0 | 6.0 | 0.0 | 0.0 |
| 13 | 14.0 | 44.0 | 75.0 | 198.0 | 0.0 | 0.0 | 0.0 | 50.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 1.0 | 0.0 | 18.0 |
| 14 | 1.0 | 1.0 | 12.0 | 0.0 | 0.0 | 0.0 | 45.0 | 0.0 | 2.0 | 2.0 | 69.0 | 2.0 | 0.0 | 0.0 | 272.0 | 0.0 |
| 15 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 3.0 | 378.0 | 0.0 | 17.0 | 1.0 | 1.0 | 0.0 | 0.0 |
| 16 | 1.0 | 1.0 | 2.0 | 0.0 | 1.0 | 6.0 | 368.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 13.0 | 0.0 |
| 17 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 8.0 | 0.0 | 3.0 | 0.0 | 335.0 | 0.0 | 48.0 | 0.0 | 10.0 | 0.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 0.0 | 232.0 | 36.0 | 57.0 | 30.0 | 0.0 | 0.0 | 0.0 | 0.0 | 26.0 | 4.0 | 17.0 | 4.0 |
| 19 | 56.0 | 68.0 | 112.0 | 22.0 | 14.0 | 9.0 | 5.0 | 12.0 | 17.0 | 26.0 | 24.0 | 7.0 | 10.0 | 13.0 | 7.0 | 4.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.165 | 0.229 | 0.061 | -0.733 | -0.186 | 0.567 | -0.733 | -0.097 | 0.198 | 0.631 | -0.337 | 0.024 | -0.453 | 0.024 | -0.516 | -0.055 |
| 02 | -0.453 | -0.015 | 0.61 | -0.909 | 0.373 | 0.631 | 0.318 | 0.198 | -0.14 | -0.393 | -0.337 | -0.516 | -0.733 | 0.198 | 0.229 | -0.909 |
| 03 | -0.097 | 0.061 | 0.061 | -0.909 | -0.655 | 0.345 | 0.198 | 0.474 | 0.061 | 0.544 | 0.521 | -0.337 | -1.25 | -0.015 | -0.097 | -0.817 |
| 04 | -0.733 | -0.055 | -0.186 | -0.655 | -0.337 | 0.766 | -0.583 | 0.567 | -0.015 | 0.916 | -0.284 | -0.453 | -2.032 | 0.544 | -0.337 | -0.516 |
| 05 | -1.772 | 0.132 | -0.14 | -2.032 | 0.947 | 0.61 | -0.393 | 0.801 | -2.032 | 0.425 | -0.337 | -0.733 | -0.733 | 0.691 | -0.14 | -1.772 |
| 06 | -0.393 | 0.45 | -0.055 | -1.772 | 0.498 | 1.061 | -0.909 | 0.45 | -0.583 | 0.289 | -0.337 | -0.655 | -1.772 | 0.801 | -0.909 | -1.567 |
| 07 | -1.01 | 0.024 | -0.516 | 0.061 | -1.396 | 0.691 | -0.516 | 1.645 | -1.25 | -1.01 | -1.123 | 0.399 | -2.032 | -0.393 | -0.453 | 0.024 |
| 08 | -2.93 | -4.228 | -0.097 | -2.93 | 0.024 | -2.032 | 1.061 | -4.228 | -1.01 | -2.383 | 0.521 | -4.228 | -0.234 | -2.93 | 2.067 | -4.228 |
| 09 | -2.032 | 0.691 | -2.383 | -4.228 | -4.228 | -1.396 | -4.228 | -4.228 | 0.399 | 2.457 | -1.567 | -4.228 | -4.228 | -2.93 | -4.228 | -4.228 |
| 10 | -4.228 | -4.228 | 0.474 | -4.228 | -0.516 | -2.383 | 2.582 | -2.93 | -2.383 | -4.228 | -1.567 | -4.228 | -4.228 | -4.228 | -4.228 | -4.228 |
| 11 | -4.228 | -0.516 | -2.383 | -4.228 | -4.228 | -2.93 | -4.228 | -2.93 | -2.93 | 2.685 | -2.032 | -1.567 | -4.228 | -2.93 | -4.228 | -4.228 |
| 12 | -4.228 | -4.228 | -4.228 | -2.93 | 2.552 | 0.474 | -1.396 | -0.393 | -2.93 | -2.032 | -4.228 | -2.93 | -4.228 | -1.396 | -4.228 | -4.228 |
| 13 | -0.583 | 0.544 | 1.074 | 2.042 | -4.228 | -4.228 | -4.228 | 0.671 | -4.228 | -4.228 | -4.228 | -1.396 | -4.228 | -2.93 | -4.228 | -0.337 |
| 14 | -2.93 | -2.93 | -0.733 | -4.228 | -4.228 | -4.228 | 0.567 | -4.228 | -2.383 | -2.383 | 0.991 | -2.383 | -4.228 | -4.228 | 2.359 | -4.228 |
| 15 | -4.228 | -2.032 | -4.228 | -4.228 | -4.228 | -2.032 | -4.228 | -4.228 | -2.032 | 2.687 | -4.228 | -0.393 | -2.93 | -2.93 | -4.228 | -4.228 |
| 16 | -2.93 | -2.93 | -2.383 | -4.228 | -2.93 | -1.396 | 2.661 | -0.909 | -4.228 | -4.228 | -4.228 | -4.228 | -4.228 | -1.772 | -0.655 | -4.228 |
| 17 | -4.228 | -2.383 | -4.228 | -4.228 | -4.228 | -1.123 | -4.228 | -2.032 | -4.228 | 2.567 | -4.228 | 0.631 | -4.228 | -0.909 | -4.228 | -4.228 |
| 18 | -4.228 | -4.228 | -4.228 | -4.228 | 2.2 | 0.345 | 0.801 | 0.165 | -4.228 | -4.228 | -4.228 | -4.228 | 0.024 | -1.772 | -0.393 | -1.772 |
| 19 | 0.784 | 0.977 | 1.473 | -0.14 | -0.583 | -1.01 | -1.567 | -0.733 | -0.393 | 0.024 | -0.055 | -1.25 | -0.909 | -0.655 | -1.25 | -1.772 |