Model info
| Transcription factor | Ovol2 | ||||||||
| Model | OVOL2_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 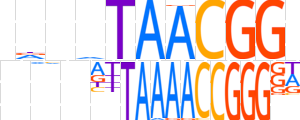 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 11 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndndTAACGGd | ||||||||
| Best auROC (human) | 0.454 | ||||||||
| Best auROC (mouse) | 0.963 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 185 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | OVOL-factors {2.3.3.17} | ||||||||
| MGI | MGI:1338039 | ||||||||
| EntrezGene | GeneID:107586 (SSTAR profile) | ||||||||
| UniProt ID | OVOL2_MOUSE | ||||||||
| UniProt AC | Q8CIV7 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Ovol2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 27.0 | 4.0 | 5.0 | 7.0 | 23.0 | 5.0 | 14.0 | 4.0 | 30.0 | 5.0 | 13.0 | 5.0 | 14.0 | 4.0 | 14.0 | 11.0 |
| 02 | 29.0 | 29.0 | 25.0 | 11.0 | 5.0 | 6.0 | 0.0 | 7.0 | 19.0 | 9.0 | 11.0 | 7.0 | 3.0 | 10.0 | 1.0 | 13.0 |
| 03 | 31.0 | 4.0 | 18.0 | 3.0 | 18.0 | 13.0 | 6.0 | 17.0 | 15.0 | 6.0 | 8.0 | 8.0 | 16.0 | 7.0 | 11.0 | 4.0 |
| 04 | 0.0 | 0.0 | 0.0 | 80.0 | 0.0 | 0.0 | 0.0 | 30.0 | 0.0 | 0.0 | 0.0 | 43.0 | 0.0 | 0.0 | 0.0 | 32.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 185.0 | 0.0 | 0.0 | 0.0 |
| 06 | 180.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 180.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 185.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 185.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 67.0 | 10.0 | 39.0 | 69.0 | 0.0 | 0.0 | 0.0 | 0.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.832 | -1.011 | -0.803 | -0.484 | 0.674 | -0.803 | 0.187 | -1.011 | 0.936 | -0.803 | 0.114 | -0.803 | 0.187 | -1.011 | 0.187 | -0.048 |
| 02 | 0.903 | 0.903 | 0.756 | -0.048 | -0.803 | -0.631 | -3.596 | -0.484 | 0.486 | -0.243 | -0.048 | -0.484 | -1.274 | -0.141 | -2.193 | 0.114 |
| 03 | 0.969 | -1.011 | 0.433 | -1.274 | 0.433 | 0.114 | -0.631 | 0.377 | 0.254 | -0.631 | -0.356 | -0.356 | 0.317 | -0.484 | -0.048 | -1.011 |
| 04 | -3.596 | -3.596 | -3.596 | 1.911 | -3.596 | -3.596 | -3.596 | 0.936 | -3.596 | -3.596 | -3.596 | 1.293 | -3.596 | -3.596 | -3.596 | 1.0 |
| 05 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | 2.747 | -3.596 | -3.596 | -3.596 |
| 06 | 2.719 | -3.596 | -0.803 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 |
| 07 | -3.596 | 2.719 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -0.803 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 |
| 08 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | 2.747 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 |
| 09 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | 2.747 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 |
| 10 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | -3.596 | 1.734 | -0.141 | 1.196 | 1.763 | -3.596 | -3.596 | -3.596 | -3.596 |