Model info
| Transcription factor | TP53 (GeneCards) | ||||||||
| Model | P53_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 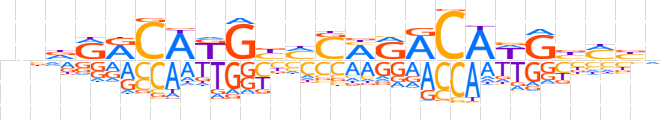 | ||||||||
| LOGO (reverse complement) | 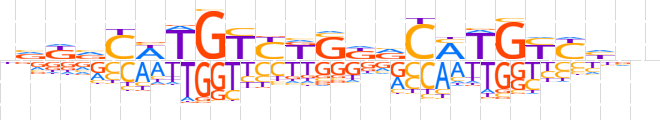 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nndRRCAKGbMSMRACAWGbMbn | ||||||||
| Best auROC (human) | 0.997 | ||||||||
| Best auROC (mouse) | 0.988 | ||||||||
| Peak sets in benchmark (human) | 141 | ||||||||
| Peak sets in benchmark (mouse) | 39 | ||||||||
| Aligned words | 598 | ||||||||
| TF family | p53-related factors {6.3.1} | ||||||||
| TF subfamily | p53 {6.3.1.0.1} | ||||||||
| HGNC | HGNC:11998 | ||||||||
| EntrezGene | GeneID:7157 (SSTAR profile) | ||||||||
| UniProt ID | P53_HUMAN | ||||||||
| UniProt AC | P04637 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | TP53 expression | ||||||||
| ReMap ChIP-seq dataset list | TP53 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 17.0 | 68.0 | 65.0 | 15.0 | 18.0 | 41.0 | 2.0 | 32.0 | 22.0 | 36.0 | 47.0 | 40.0 | 14.0 | 21.0 | 39.0 | 12.0 |
| 02 | 18.0 | 8.0 | 38.0 | 7.0 | 114.0 | 9.0 | 17.0 | 26.0 | 69.0 | 7.0 | 34.0 | 43.0 | 24.0 | 2.0 | 64.0 | 9.0 |
| 03 | 24.0 | 9.0 | 175.0 | 17.0 | 15.0 | 1.0 | 2.0 | 8.0 | 30.0 | 3.0 | 110.0 | 10.0 | 8.0 | 1.0 | 61.0 | 15.0 |
| 04 | 64.0 | 1.0 | 9.0 | 3.0 | 12.0 | 0.0 | 2.0 | 0.0 | 193.0 | 4.0 | 139.0 | 12.0 | 29.0 | 1.0 | 11.0 | 9.0 |
| 05 | 0.0 | 246.0 | 47.0 | 5.0 | 0.0 | 5.0 | 0.0 | 1.0 | 0.0 | 160.0 | 0.0 | 1.0 | 0.0 | 20.0 | 2.0 | 2.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 349.0 | 16.0 | 14.0 | 52.0 | 47.0 | 1.0 | 0.0 | 1.0 | 1.0 | 4.0 | 3.0 | 1.0 |
| 07 | 37.0 | 11.0 | 75.0 | 274.0 | 1.0 | 1.0 | 0.0 | 19.0 | 2.0 | 0.0 | 3.0 | 12.0 | 11.0 | 1.0 | 1.0 | 41.0 |
| 08 | 0.0 | 0.0 | 50.0 | 1.0 | 6.0 | 0.0 | 7.0 | 0.0 | 63.0 | 0.0 | 16.0 | 0.0 | 1.0 | 0.0 | 344.0 | 1.0 |
| 09 | 1.0 | 9.0 | 59.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 21.0 | 223.0 | 1.0 | 172.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 10 | 6.0 | 9.0 | 5.0 | 2.0 | 27.0 | 180.0 | 5.0 | 20.0 | 58.0 | 2.0 | 0.0 | 0.0 | 19.0 | 95.0 | 23.0 | 38.0 |
| 11 | 2.0 | 56.0 | 49.0 | 3.0 | 9.0 | 257.0 | 3.0 | 17.0 | 3.0 | 27.0 | 2.0 | 1.0 | 1.0 | 42.0 | 9.0 | 8.0 |
| 12 | 7.0 | 1.0 | 6.0 | 1.0 | 252.0 | 59.0 | 32.0 | 39.0 | 58.0 | 1.0 | 1.0 | 3.0 | 26.0 | 1.0 | 0.0 | 2.0 |
| 13 | 45.0 | 17.0 | 273.0 | 8.0 | 51.0 | 4.0 | 3.0 | 4.0 | 4.0 | 0.0 | 33.0 | 2.0 | 4.0 | 2.0 | 35.0 | 4.0 |
| 14 | 85.0 | 1.0 | 16.0 | 2.0 | 23.0 | 0.0 | 0.0 | 0.0 | 236.0 | 0.0 | 107.0 | 1.0 | 8.0 | 0.0 | 5.0 | 5.0 |
| 15 | 2.0 | 335.0 | 13.0 | 2.0 | 0.0 | 1.0 | 0.0 | 0.0 | 5.0 | 122.0 | 1.0 | 0.0 | 0.0 | 7.0 | 1.0 | 0.0 |
| 16 | 0.0 | 7.0 | 0.0 | 0.0 | 395.0 | 12.0 | 8.0 | 50.0 | 13.0 | 1.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| 17 | 56.0 | 27.0 | 58.0 | 268.0 | 7.0 | 2.0 | 0.0 | 11.0 | 2.0 | 2.0 | 1.0 | 4.0 | 15.0 | 1.0 | 1.0 | 34.0 |
| 18 | 10.0 | 2.0 | 56.0 | 12.0 | 16.0 | 3.0 | 13.0 | 0.0 | 56.0 | 0.0 | 3.0 | 1.0 | 4.0 | 3.0 | 309.0 | 1.0 |
| 19 | 3.0 | 15.0 | 60.0 | 8.0 | 2.0 | 5.0 | 1.0 | 0.0 | 20.0 | 207.0 | 11.0 | 143.0 | 0.0 | 0.0 | 10.0 | 4.0 |
| 20 | 7.0 | 12.0 | 1.0 | 5.0 | 43.0 | 165.0 | 2.0 | 17.0 | 58.0 | 19.0 | 1.0 | 4.0 | 17.0 | 82.0 | 7.0 | 49.0 |
| 21 | 5.0 | 43.0 | 66.0 | 11.0 | 25.0 | 154.0 | 6.0 | 93.0 | 0.0 | 11.0 | 0.0 | 0.0 | 2.0 | 44.0 | 11.0 | 18.0 |
| 22 | 6.0 | 13.0 | 9.0 | 4.0 | 110.0 | 57.0 | 30.0 | 55.0 | 54.0 | 13.0 | 12.0 | 4.0 | 17.0 | 20.0 | 51.0 | 34.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.577 | 0.793 | 0.748 | -0.699 | -0.521 | 0.291 | -2.562 | 0.045 | -0.324 | 0.162 | 0.426 | 0.266 | -0.766 | -0.37 | 0.241 | -0.916 |
| 02 | -0.521 | -1.306 | 0.215 | -1.433 | 1.307 | -1.193 | -0.577 | -0.159 | 0.807 | -1.433 | 0.105 | 0.338 | -0.238 | -2.562 | 0.733 | -1.193 |
| 03 | -0.238 | -1.193 | 1.735 | -0.577 | -0.699 | -3.105 | -2.562 | -1.306 | -0.018 | -2.212 | 1.272 | -1.092 | -1.306 | -3.105 | 0.685 | -0.699 |
| 04 | 0.733 | -3.105 | -1.193 | -2.212 | -0.916 | -4.382 | -2.562 | -4.382 | 1.832 | -1.954 | 1.505 | -0.916 | -0.052 | -3.105 | -1.0 | -1.193 |
| 05 | -4.382 | 2.075 | 0.426 | -1.748 | -4.382 | -1.748 | -4.382 | -3.105 | -4.382 | 1.645 | -4.382 | -3.105 | -4.382 | -0.417 | -2.562 | -2.562 |
| 06 | -4.382 | -4.382 | -4.382 | -4.382 | 2.424 | -0.636 | -0.766 | 0.526 | 0.426 | -3.105 | -4.382 | -3.105 | -3.105 | -1.954 | -2.212 | -3.105 |
| 07 | 0.189 | -1.0 | 0.89 | 2.182 | -3.105 | -3.105 | -4.382 | -0.468 | -2.562 | -4.382 | -2.212 | -0.916 | -1.0 | -3.105 | -3.105 | 0.291 |
| 08 | -4.382 | -4.382 | 0.487 | -3.105 | -1.578 | -4.382 | -1.433 | -4.382 | 0.717 | -4.382 | -0.636 | -4.382 | -3.105 | -4.382 | 2.409 | -3.105 |
| 09 | -3.105 | -1.193 | 0.652 | -3.105 | -4.382 | -4.382 | -4.382 | -4.382 | -0.37 | 1.977 | -3.105 | 1.717 | -4.382 | -4.382 | -4.382 | -2.562 |
| 10 | -1.578 | -1.193 | -1.748 | -2.562 | -0.122 | 1.763 | -1.748 | -0.417 | 0.635 | -2.562 | -4.382 | -4.382 | -0.468 | 1.126 | -0.28 | 0.215 |
| 11 | -2.562 | 0.6 | 0.467 | -2.212 | -1.193 | 2.118 | -2.212 | -0.577 | -2.212 | -0.122 | -2.562 | -3.105 | -3.105 | 0.314 | -1.193 | -1.306 |
| 12 | -1.433 | -3.105 | -1.578 | -3.105 | 2.099 | 0.652 | 0.045 | 0.241 | 0.635 | -3.105 | -3.105 | -2.212 | -0.159 | -3.105 | -4.382 | -2.562 |
| 13 | 0.383 | -0.577 | 2.179 | -1.306 | 0.507 | -1.954 | -2.212 | -1.954 | -1.954 | -4.382 | 0.076 | -2.562 | -1.954 | -2.562 | 0.134 | -1.954 |
| 14 | 1.015 | -3.105 | -0.636 | -2.562 | -0.28 | -4.382 | -4.382 | -4.382 | 2.033 | -4.382 | 1.244 | -3.105 | -1.306 | -4.382 | -1.748 | -1.748 |
| 15 | -2.562 | 2.383 | -0.838 | -2.562 | -4.382 | -3.105 | -4.382 | -4.382 | -1.748 | 1.375 | -3.105 | -4.382 | -4.382 | -1.433 | -3.105 | -4.382 |
| 16 | -4.382 | -1.433 | -4.382 | -4.382 | 2.548 | -0.916 | -1.306 | 0.487 | -0.838 | -3.105 | -4.382 | -3.105 | -3.105 | -4.382 | -3.105 | -4.382 |
| 17 | 0.6 | -0.122 | 0.635 | 2.16 | -1.433 | -2.562 | -4.382 | -1.0 | -2.562 | -2.562 | -3.105 | -1.954 | -0.699 | -3.105 | -3.105 | 0.105 |
| 18 | -1.092 | -2.562 | 0.6 | -0.916 | -0.636 | -2.212 | -0.838 | -4.382 | 0.6 | -4.382 | -2.212 | -3.105 | -1.954 | -2.212 | 2.302 | -3.105 |
| 19 | -2.212 | -0.699 | 0.668 | -1.306 | -2.562 | -1.748 | -3.105 | -4.382 | -0.417 | 1.902 | -1.0 | 1.533 | -4.382 | -4.382 | -1.092 | -1.954 |
| 20 | -1.433 | -0.916 | -3.105 | -1.748 | 0.338 | 1.676 | -2.562 | -0.577 | 0.635 | -0.468 | -3.105 | -1.954 | -0.577 | 0.979 | -1.433 | 0.467 |
| 21 | -1.748 | 0.338 | 0.763 | -1.0 | -0.198 | 1.607 | -1.578 | 1.104 | -4.382 | -1.0 | -4.382 | -4.382 | -2.562 | 0.361 | -1.0 | -0.521 |
| 22 | -1.578 | -0.838 | -1.193 | -1.954 | 1.272 | 0.617 | -0.018 | 0.582 | 0.564 | -0.838 | -0.916 | -1.954 | -0.577 | -0.417 | 0.507 | 0.105 |