Model info
| Transcription factor | PAX2 (GeneCards) | ||||||||
| Model | PAX2_HUMAN.H11MO.0.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 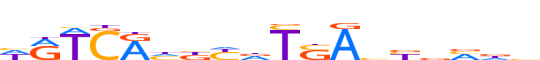 | ||||||||
| Data source | Integrative | ||||||||
| Model release | HOCOMOCOv9 | ||||||||
| Model length | 18 | ||||||||
| Quality | D | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vhbnvdTSAhRvdYGAYW | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 50 | ||||||||
| TF family | Paired domain only {3.2.2} | ||||||||
| TF subfamily | PAX-2-like factors (partial homeobox) {3.2.2.2} | ||||||||
| HGNC | HGNC:8616 | ||||||||
| EntrezGene | GeneID:5076 (SSTAR profile) | ||||||||
| UniProt ID | PAX2_HUMAN | ||||||||
| UniProt AC | Q02962 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | PAX2 expression | ||||||||
| ReMap ChIP-seq dataset list | PAX2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 11.614 | 9.182 | 20.699 | 8.734 |
| 02 | 16.679 | 13.799 | 2.879 | 16.872 |
| 03 | 2.879 | 7.865 | 12.658 | 26.826 |
| 04 | 8.91 | 23.227 | 9.182 | 8.91 |
| 05 | 27.52 | 8.041 | 10.051 | 4.617 |
| 06 | 9.779 | 8.734 | 22.806 | 8.91 |
| 07 | 0.0 | 7.769 | 0.966 | 41.494 |
| 08 | 4.02 | 29.337 | 14.861 | 2.01 |
| 09 | 41.319 | 2.01 | 6.9 | 0.0 |
| 10 | 7.593 | 12.93 | 4.714 | 24.991 |
| 11 | 10.92 | 2.976 | 29.433 | 6.9 |
| 12 | 13.202 | 26.106 | 6.031 | 4.889 |
| 13 | 9.875 | 4.617 | 25.685 | 10.051 |
| 14 | 2.879 | 12.061 | 0.0 | 35.288 |
| 15 | 4.02 | 4.889 | 41.319 | 0.0 |
| 16 | 39.212 | 0.0 | 2.01 | 9.006 |
| 17 | 3.476 | 30.671 | 0.0 | 16.082 |
| 18 | 25.863 | 2.091 | 7.253 | 15.022 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.072 | -0.287 | 0.471 | -0.332 |
| 02 | 0.266 | 0.088 | -1.255 | 0.277 |
| 03 | -1.255 | -0.426 | 0.007 | 0.72 |
| 04 | -0.314 | 0.581 | -0.287 | -0.314 |
| 05 | 0.744 | -0.406 | -0.205 | -0.883 |
| 06 | -0.23 | -0.332 | 0.564 | -0.314 |
| 07 | -2.626 | -0.437 | -1.94 | 1.144 |
| 08 | -0.996 | 0.806 | 0.157 | -1.51 |
| 09 | 1.139 | -1.51 | -0.541 | -2.626 |
| 10 | -0.457 | 0.027 | -0.866 | 0.652 |
| 11 | -0.129 | -1.23 | 0.809 | -0.541 |
| 12 | 0.047 | 0.694 | -0.658 | -0.836 |
| 13 | -0.221 | -0.883 | 0.678 | -0.205 |
| 14 | -1.255 | -0.037 | -2.626 | 0.986 |
| 15 | -0.996 | -0.836 | 1.139 | -2.626 |
| 16 | 1.088 | -2.626 | -1.51 | -0.304 |
| 17 | -1.111 | 0.849 | -2.626 | 0.231 |
| 18 | 0.685 | -1.484 | -0.497 | 0.167 |