Model info
| Transcription factor | Pax3 | ||||||||
| Model | PAX3_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 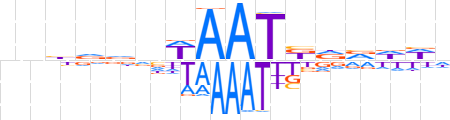 | ||||||||
| LOGO (reverse complement) | 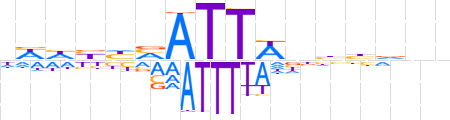 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnbvdnWAATbdvddd | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.817 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 447 | ||||||||
| TF family | Paired plus homeo domain {3.2.1} | ||||||||
| TF subfamily | PAX-3/7 {3.2.1.1} | ||||||||
| MGI | MGI:97487 | ||||||||
| EntrezGene | GeneID:18505 (SSTAR profile) | ||||||||
| UniProt ID | PAX3_MOUSE | ||||||||
| UniProt AC | P24610 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Pax3 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 15.0 | 44.0 | 34.0 | 13.0 | 69.0 | 26.0 | 25.0 | 23.0 | 12.0 | 27.0 | 31.0 | 12.0 | 15.0 | 21.0 | 65.0 | 13.0 |
| 02 | 12.0 | 25.0 | 19.0 | 55.0 | 17.0 | 39.0 | 14.0 | 48.0 | 22.0 | 36.0 | 29.0 | 68.0 | 10.0 | 12.0 | 19.0 | 20.0 |
| 03 | 25.0 | 16.0 | 13.0 | 7.0 | 52.0 | 11.0 | 30.0 | 19.0 | 30.0 | 20.0 | 17.0 | 14.0 | 23.0 | 15.0 | 145.0 | 8.0 |
| 04 | 23.0 | 17.0 | 68.0 | 22.0 | 30.0 | 14.0 | 9.0 | 9.0 | 49.0 | 26.0 | 82.0 | 48.0 | 6.0 | 3.0 | 35.0 | 4.0 |
| 05 | 14.0 | 71.0 | 16.0 | 7.0 | 24.0 | 12.0 | 3.0 | 21.0 | 84.0 | 48.0 | 29.0 | 33.0 | 6.0 | 7.0 | 68.0 | 2.0 |
| 06 | 57.0 | 1.0 | 10.0 | 60.0 | 13.0 | 5.0 | 8.0 | 112.0 | 43.0 | 4.0 | 8.0 | 61.0 | 4.0 | 2.0 | 4.0 | 53.0 |
| 07 | 113.0 | 3.0 | 1.0 | 0.0 | 11.0 | 0.0 | 0.0 | 1.0 | 28.0 | 0.0 | 1.0 | 1.0 | 277.0 | 0.0 | 8.0 | 1.0 |
| 08 | 425.0 | 0.0 | 3.0 | 1.0 | 3.0 | 0.0 | 0.0 | 0.0 | 8.0 | 2.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 |
| 09 | 10.0 | 10.0 | 6.0 | 413.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 10 | 0.0 | 3.0 | 6.0 | 1.0 | 1.0 | 1.0 | 1.0 | 7.0 | 1.0 | 1.0 | 1.0 | 3.0 | 5.0 | 93.0 | 124.0 | 197.0 |
| 11 | 1.0 | 0.0 | 5.0 | 1.0 | 55.0 | 8.0 | 21.0 | 14.0 | 56.0 | 19.0 | 37.0 | 20.0 | 16.0 | 15.0 | 161.0 | 16.0 |
| 12 | 40.0 | 12.0 | 73.0 | 3.0 | 25.0 | 7.0 | 3.0 | 7.0 | 152.0 | 31.0 | 23.0 | 18.0 | 11.0 | 15.0 | 24.0 | 1.0 |
| 13 | 30.0 | 11.0 | 14.0 | 173.0 | 30.0 | 9.0 | 3.0 | 23.0 | 54.0 | 21.0 | 19.0 | 29.0 | 4.0 | 4.0 | 12.0 | 9.0 |
| 14 | 54.0 | 7.0 | 16.0 | 41.0 | 13.0 | 11.0 | 2.0 | 19.0 | 22.0 | 6.0 | 9.0 | 11.0 | 12.0 | 16.0 | 19.0 | 187.0 |
| 15 | 60.0 | 5.0 | 11.0 | 25.0 | 21.0 | 8.0 | 4.0 | 7.0 | 16.0 | 10.0 | 11.0 | 9.0 | 127.0 | 22.0 | 29.0 | 80.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.606 | 0.454 | 0.198 | -0.745 | 0.901 | -0.066 | -0.105 | -0.187 | -0.823 | -0.029 | 0.107 | -0.823 | -0.606 | -0.277 | 0.841 | -0.745 |
| 02 | -0.823 | -0.105 | -0.375 | 0.675 | -0.484 | 0.334 | -0.673 | 0.54 | -0.231 | 0.255 | 0.041 | 0.886 | -0.999 | -0.823 | -0.375 | -0.324 |
| 03 | -0.105 | -0.543 | -0.745 | -1.34 | 0.619 | -0.907 | 0.075 | -0.375 | 0.075 | -0.324 | -0.484 | -0.673 | -0.187 | -0.606 | 1.64 | -1.213 |
| 04 | -0.187 | -0.484 | 0.886 | -0.231 | 0.075 | -0.673 | -1.1 | -1.1 | 0.56 | -0.066 | 1.072 | 0.54 | -1.486 | -2.121 | 0.227 | -1.862 |
| 05 | -0.673 | 0.929 | -0.543 | -1.34 | -0.145 | -0.823 | -2.121 | -0.277 | 1.096 | 0.54 | 0.041 | 0.169 | -1.486 | -1.34 | 0.886 | -2.472 |
| 06 | 0.711 | -3.016 | -0.999 | 0.762 | -0.745 | -1.656 | -1.213 | 1.383 | 0.431 | -1.862 | -1.213 | 0.778 | -1.862 | -2.472 | -1.862 | 0.638 |
| 07 | 1.392 | -2.121 | -3.016 | -4.304 | -0.907 | -4.304 | -4.304 | -3.016 | 0.007 | -4.304 | -3.016 | -3.016 | 2.286 | -4.304 | -1.213 | -3.016 |
| 08 | 2.714 | -4.304 | -2.121 | -3.016 | -2.121 | -4.304 | -4.304 | -4.304 | -1.213 | -2.472 | -4.304 | -4.304 | -2.121 | -4.304 | -4.304 | -4.304 |
| 09 | -0.999 | -0.999 | -1.486 | 2.685 | -4.304 | -4.304 | -4.304 | -2.472 | -4.304 | -4.304 | -4.304 | -2.121 | -4.304 | -4.304 | -4.304 | -3.016 |
| 10 | -4.304 | -2.121 | -1.486 | -3.016 | -3.016 | -3.016 | -3.016 | -1.34 | -3.016 | -3.016 | -3.016 | -2.121 | -1.656 | 1.198 | 1.484 | 1.946 |
| 11 | -3.016 | -4.304 | -1.656 | -3.016 | 0.675 | -1.213 | -0.277 | -0.673 | 0.693 | -0.375 | 0.282 | -0.324 | -0.543 | -0.606 | 1.745 | -0.543 |
| 12 | 0.359 | -0.823 | 0.957 | -2.121 | -0.105 | -1.34 | -2.121 | -1.34 | 1.687 | 0.107 | -0.187 | -0.428 | -0.907 | -0.606 | -0.145 | -3.016 |
| 13 | 0.075 | -0.907 | -0.673 | 1.816 | 0.075 | -1.1 | -2.121 | -0.187 | 0.657 | -0.277 | -0.375 | 0.041 | -1.862 | -1.862 | -0.823 | -1.1 |
| 14 | 0.657 | -1.34 | -0.543 | 0.384 | -0.745 | -0.907 | -2.472 | -0.375 | -0.231 | -1.486 | -1.1 | -0.907 | -0.823 | -0.543 | -0.375 | 1.894 |
| 15 | 0.762 | -1.656 | -0.907 | -0.105 | -0.277 | -1.213 | -1.862 | -1.34 | -0.543 | -0.999 | -0.907 | -1.1 | 1.508 | -0.231 | 0.041 | 1.048 |