Model info
| Transcription factor | Pax5 | ||||||||
| Model | PAX5_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 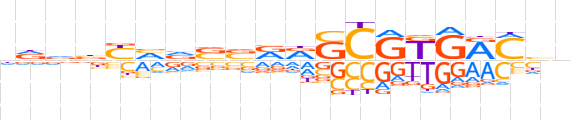 | ||||||||
| LOGO (reverse complement) | 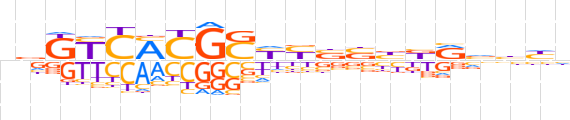 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nddbYvvvvRRSCRKGRMbn | ||||||||
| Best auROC (human) | 0.947 | ||||||||
| Best auROC (mouse) | 0.927 | ||||||||
| Peak sets in benchmark (human) | 32 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 401 | ||||||||
| TF family | Paired domain only {3.2.2} | ||||||||
| TF subfamily | PAX-2-like factors (partial homeobox) {3.2.2.2} | ||||||||
| MGI | MGI:97489 | ||||||||
| EntrezGene | GeneID:18507 (SSTAR profile) | ||||||||
| UniProt ID | PAX5_MOUSE | ||||||||
| UniProt AC | Q02650 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Pax5 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 55.0 | 3.0 | 37.0 | 13.0 | 24.0 | 2.0 | 22.0 | 11.0 | 38.0 | 14.0 | 68.0 | 21.0 | 5.0 | 3.0 | 68.0 | 8.0 |
| 02 | 48.0 | 19.0 | 44.0 | 11.0 | 7.0 | 4.0 | 5.0 | 6.0 | 42.0 | 33.0 | 76.0 | 44.0 | 7.0 | 1.0 | 41.0 | 4.0 |
| 03 | 15.0 | 32.0 | 41.0 | 16.0 | 2.0 | 18.0 | 9.0 | 28.0 | 17.0 | 39.0 | 74.0 | 36.0 | 0.0 | 10.0 | 43.0 | 12.0 |
| 04 | 3.0 | 14.0 | 8.0 | 9.0 | 2.0 | 57.0 | 10.0 | 30.0 | 6.0 | 110.0 | 21.0 | 30.0 | 3.0 | 67.0 | 12.0 | 10.0 |
| 05 | 5.0 | 3.0 | 3.0 | 3.0 | 181.0 | 32.0 | 24.0 | 11.0 | 1.0 | 44.0 | 3.0 | 3.0 | 15.0 | 41.0 | 14.0 | 9.0 |
| 06 | 27.0 | 26.0 | 132.0 | 17.0 | 99.0 | 7.0 | 13.0 | 1.0 | 5.0 | 17.0 | 16.0 | 6.0 | 17.0 | 7.0 | 2.0 | 0.0 |
| 07 | 23.0 | 38.0 | 78.0 | 9.0 | 14.0 | 31.0 | 9.0 | 3.0 | 3.0 | 112.0 | 40.0 | 8.0 | 0.0 | 7.0 | 17.0 | 0.0 |
| 08 | 3.0 | 11.0 | 23.0 | 3.0 | 19.0 | 114.0 | 45.0 | 10.0 | 22.0 | 70.0 | 38.0 | 14.0 | 0.0 | 8.0 | 11.0 | 1.0 |
| 09 | 10.0 | 4.0 | 29.0 | 1.0 | 134.0 | 22.0 | 38.0 | 9.0 | 33.0 | 11.0 | 70.0 | 3.0 | 8.0 | 2.0 | 18.0 | 0.0 |
| 10 | 98.0 | 8.0 | 39.0 | 40.0 | 29.0 | 2.0 | 2.0 | 6.0 | 93.0 | 6.0 | 40.0 | 16.0 | 6.0 | 1.0 | 1.0 | 5.0 |
| 11 | 0.0 | 104.0 | 121.0 | 1.0 | 0.0 | 11.0 | 6.0 | 0.0 | 1.0 | 19.0 | 61.0 | 1.0 | 0.0 | 21.0 | 46.0 | 0.0 |
| 12 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 138.0 | 0.0 | 17.0 | 0.0 | 184.0 | 6.0 | 44.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 13 | 0.0 | 0.0 | 0.0 | 0.0 | 82.0 | 5.0 | 226.0 | 9.0 | 2.0 | 0.0 | 4.0 | 0.0 | 5.0 | 0.0 | 59.0 | 0.0 |
| 14 | 7.0 | 2.0 | 12.0 | 68.0 | 0.0 | 1.0 | 0.0 | 4.0 | 10.0 | 16.0 | 56.0 | 207.0 | 0.0 | 1.0 | 1.0 | 7.0 |
| 15 | 3.0 | 1.0 | 13.0 | 0.0 | 6.0 | 0.0 | 14.0 | 0.0 | 63.0 | 1.0 | 5.0 | 0.0 | 25.0 | 1.0 | 257.0 | 3.0 |
| 16 | 66.0 | 1.0 | 23.0 | 7.0 | 2.0 | 0.0 | 1.0 | 0.0 | 212.0 | 21.0 | 44.0 | 12.0 | 1.0 | 0.0 | 1.0 | 1.0 |
| 17 | 34.0 | 240.0 | 4.0 | 3.0 | 10.0 | 4.0 | 1.0 | 7.0 | 17.0 | 41.0 | 1.0 | 10.0 | 2.0 | 11.0 | 2.0 | 5.0 |
| 18 | 13.0 | 18.0 | 20.0 | 12.0 | 48.0 | 145.0 | 49.0 | 54.0 | 3.0 | 3.0 | 0.0 | 2.0 | 3.0 | 4.0 | 3.0 | 15.0 |
| 19 | 25.0 | 12.0 | 24.0 | 6.0 | 68.0 | 32.0 | 24.0 | 46.0 | 12.0 | 28.0 | 21.0 | 11.0 | 13.0 | 43.0 | 17.0 | 10.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.8 | -1.998 | 0.407 | -0.621 | -0.02 | -2.35 | -0.106 | -0.783 | 0.434 | -0.548 | 1.011 | -0.152 | -1.532 | -1.998 | 1.011 | -1.089 |
| 02 | 0.665 | -0.25 | 0.579 | -0.783 | -1.216 | -1.738 | -1.532 | -1.362 | 0.533 | 0.294 | 1.122 | 0.579 | -1.216 | -2.897 | 0.509 | -1.738 |
| 03 | -0.481 | 0.264 | 0.509 | -0.418 | -2.35 | -0.303 | -0.976 | 0.132 | -0.359 | 0.459 | 1.095 | 0.38 | -4.199 | -0.875 | 0.556 | -0.698 |
| 04 | -1.998 | -0.548 | -1.089 | -0.976 | -2.35 | 0.836 | -0.875 | 0.2 | -1.362 | 1.49 | -0.152 | 0.2 | -1.998 | 0.996 | -0.698 | -0.875 |
| 05 | -1.532 | -1.998 | -1.998 | -1.998 | 1.987 | 0.264 | -0.02 | -0.783 | -2.897 | 0.579 | -1.998 | -1.998 | -0.481 | 0.509 | -0.548 | -0.976 |
| 06 | 0.096 | 0.059 | 1.672 | -0.359 | 1.385 | -1.216 | -0.621 | -2.897 | -1.532 | -0.359 | -0.418 | -1.362 | -0.359 | -1.216 | -2.35 | -4.199 |
| 07 | -0.062 | 0.434 | 1.148 | -0.976 | -0.548 | 0.232 | -0.976 | -1.998 | -1.998 | 1.508 | 0.484 | -1.089 | -4.199 | -1.216 | -0.359 | -4.199 |
| 08 | -1.998 | -0.783 | -0.062 | -1.998 | -0.25 | 1.526 | 0.601 | -0.875 | -0.106 | 1.04 | 0.434 | -0.548 | -4.199 | -1.089 | -0.783 | -2.897 |
| 09 | -0.875 | -1.738 | 0.166 | -2.897 | 1.687 | -0.106 | 0.434 | -0.976 | 0.294 | -0.783 | 1.04 | -1.998 | -1.089 | -2.35 | -0.303 | -4.199 |
| 10 | 1.375 | -1.089 | 0.459 | 0.484 | 0.166 | -2.35 | -2.35 | -1.362 | 1.323 | -1.362 | 0.484 | -0.418 | -1.362 | -2.897 | -2.897 | -1.532 |
| 11 | -4.199 | 1.434 | 1.585 | -2.897 | -4.199 | -0.783 | -1.362 | -4.199 | -2.897 | -0.25 | 0.903 | -2.897 | -4.199 | -0.152 | 0.623 | -4.199 |
| 12 | -4.199 | -4.199 | -4.199 | -2.897 | -4.199 | 1.716 | -4.199 | -0.359 | -4.199 | 2.003 | -1.362 | 0.579 | -4.199 | -4.199 | -4.199 | -2.35 |
| 13 | -4.199 | -4.199 | -4.199 | -4.199 | 1.197 | -1.532 | 2.208 | -0.976 | -2.35 | -4.199 | -1.738 | -4.199 | -1.532 | -4.199 | 0.87 | -4.199 |
| 14 | -1.216 | -2.35 | -0.698 | 1.011 | -4.199 | -2.897 | -4.199 | -1.738 | -0.875 | -0.418 | 0.818 | 2.121 | -4.199 | -2.897 | -2.897 | -1.216 |
| 15 | -1.998 | -2.897 | -0.621 | -4.199 | -1.362 | -4.199 | -0.548 | -4.199 | 0.935 | -2.897 | -1.532 | -4.199 | 0.02 | -2.897 | 2.337 | -1.998 |
| 16 | 0.982 | -2.897 | -0.062 | -1.216 | -2.35 | -4.199 | -2.897 | -4.199 | 2.145 | -0.152 | 0.579 | -0.698 | -2.897 | -4.199 | -2.897 | -2.897 |
| 17 | 0.323 | 2.268 | -1.738 | -1.998 | -0.875 | -1.738 | -2.897 | -1.216 | -0.359 | 0.509 | -2.897 | -0.875 | -2.35 | -0.783 | -2.35 | -1.532 |
| 18 | -0.621 | -0.303 | -0.2 | -0.698 | 0.665 | 1.766 | 0.686 | 0.782 | -1.998 | -1.998 | -4.199 | -2.35 | -1.998 | -1.738 | -1.998 | -0.481 |
| 19 | 0.02 | -0.698 | -0.02 | -1.362 | 1.011 | 0.264 | -0.02 | 0.623 | -0.698 | 0.132 | -0.152 | -0.783 | -0.621 | 0.556 | -0.359 | -0.875 |