Model info
| Transcription factor | Pax6 | ||||||||
| Model | PAX6_MOUSE.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 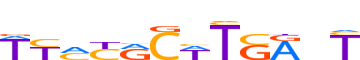 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | WYMYRCWTSRnT | ||||||||
| Best auROC (human) | 0.749 | ||||||||
| Best auROC (mouse) | 0.734 | ||||||||
| Peak sets in benchmark (human) | 6 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Paired plus homeo domain {3.2.1} | ||||||||
| TF subfamily | PAX-4/6 {3.2.1.2} | ||||||||
| MGI | MGI:97490 | ||||||||
| EntrezGene | GeneID:18508 (SSTAR profile) | ||||||||
| UniProt ID | PAX6_MOUSE | ||||||||
| UniProt AC | P63015 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Pax6 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 62.0 | 16.0 | 37.0 | 385.0 |
| 02 | 11.0 | 171.0 | 19.0 | 299.0 |
| 03 | 208.0 | 234.0 | 25.0 | 33.0 |
| 04 | 22.0 | 283.0 | 20.0 | 175.0 |
| 05 | 143.0 | 35.0 | 303.0 | 19.0 |
| 06 | 5.0 | 402.0 | 83.0 | 10.0 |
| 07 | 129.0 | 68.0 | 9.0 | 294.0 |
| 08 | 14.0 | 38.0 | 9.0 | 439.0 |
| 09 | 7.0 | 204.0 | 284.0 | 5.0 |
| 10 | 355.0 | 8.0 | 118.0 | 19.0 |
| 11 | 73.0 | 179.0 | 144.0 | 104.0 |
| 12 | 33.0 | 47.0 | 16.0 | 404.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.689 | -1.975 | -1.189 | 1.117 |
| 02 | -2.311 | 0.31 | -1.818 | 0.865 |
| 03 | 0.504 | 0.621 | -1.561 | -1.298 |
| 04 | -1.681 | 0.81 | -1.77 | 0.333 |
| 05 | 0.133 | -1.242 | 0.878 | -1.818 |
| 06 | -2.961 | 1.16 | -0.403 | -2.394 |
| 07 | 0.031 | -0.599 | -2.484 | 0.848 |
| 08 | -2.096 | -1.163 | -2.484 | 1.247 |
| 09 | -2.694 | 0.485 | 0.814 | -2.961 |
| 10 | 1.036 | -2.584 | -0.057 | -1.818 |
| 11 | -0.529 | 0.355 | 0.14 | -0.181 |
| 12 | -1.298 | -0.958 | -1.975 | 1.165 |