Model info
| Transcription factor | PBX3 (GeneCards) | ||||||||
| Model | PBX3_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 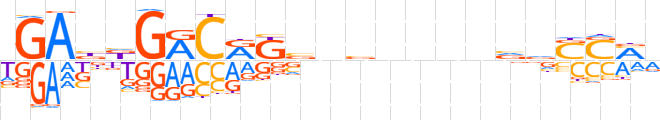 | ||||||||
| LOGO (reverse complement) | 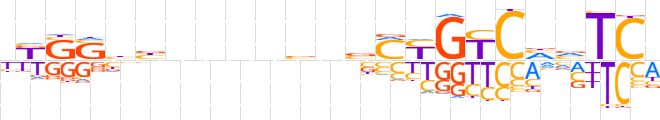 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dGAbdGRCRKbnvnnnnvvSSMv | ||||||||
| Best auROC (human) | 0.98 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 17 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 504 | ||||||||
| TF family | TALE-type homeo domain factors {3.1.4} | ||||||||
| TF subfamily | PBX {3.1.4.4} | ||||||||
| HGNC | HGNC:8634 | ||||||||
| EntrezGene | GeneID:5090 (SSTAR profile) | ||||||||
| UniProt ID | PBX3_HUMAN | ||||||||
| UniProt AC | P40426 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | PBX3 expression | ||||||||
| ReMap ChIP-seq dataset list | PBX3 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 6.201 | 0.798 | 71.543 | 0.0 | 1.01 | 1.672 | 52.586 | 2.898 | 11.71 | 9.603 | 64.929 | 1.125 | 2.038 | 1.702 | 271.226 | 0.96 |
| 02 | 14.262 | 1.74 | 3.056 | 1.901 | 11.987 | 0.798 | 0.0 | 0.99 | 439.898 | 3.699 | 16.687 | 0.0 | 3.845 | 0.0 | 1.138 | 0.0 |
| 03 | 14.19 | 122.726 | 165.075 | 168.001 | 0.0 | 2.04 | 2.656 | 1.541 | 0.0 | 8.142 | 9.693 | 3.045 | 0.0 | 0.0 | 2.891 | 0.0 |
| 04 | 2.823 | 2.081 | 3.911 | 5.375 | 19.504 | 9.177 | 49.063 | 55.165 | 17.457 | 35.876 | 15.294 | 111.687 | 11.438 | 3.955 | 53.839 | 103.356 |
| 05 | 3.566 | 0.0 | 47.656 | 0.0 | 3.332 | 1.022 | 46.734 | 0.0 | 12.471 | 0.0 | 109.636 | 0.0 | 2.017 | 0.0 | 271.716 | 1.85 |
| 06 | 12.979 | 0.0 | 8.407 | 0.0 | 0.0 | 0.0 | 1.022 | 0.0 | 312.223 | 1.857 | 158.739 | 2.922 | 0.0 | 0.0 | 0.0 | 1.85 |
| 07 | 2.772 | 291.972 | 6.229 | 24.229 | 0.0 | 0.756 | 1.1 | 0.0 | 0.0 | 161.427 | 1.648 | 5.094 | 0.0 | 4.772 | 0.0 | 0.0 |
| 08 | 1.865 | 0.0 | 0.907 | 0.0 | 271.695 | 3.994 | 157.881 | 25.358 | 1.677 | 2.03 | 4.362 | 0.907 | 7.709 | 0.0 | 15.897 | 5.717 |
| 09 | 8.454 | 23.206 | 235.617 | 15.669 | 0.0 | 1.947 | 3.144 | 0.933 | 9.058 | 18.998 | 121.576 | 29.415 | 0.99 | 6.607 | 16.184 | 8.201 |
| 10 | 1.536 | 5.432 | 10.74 | 0.794 | 6.645 | 10.103 | 15.458 | 18.553 | 44.404 | 150.178 | 153.976 | 27.963 | 1.094 | 16.955 | 29.461 | 6.709 |
| 11 | 17.573 | 11.194 | 16.197 | 8.715 | 25.976 | 68.881 | 42.328 | 45.482 | 50.029 | 87.935 | 45.14 | 26.532 | 3.812 | 15.571 | 19.543 | 15.093 |
| 12 | 14.097 | 18.219 | 49.885 | 15.189 | 41.222 | 52.866 | 64.275 | 25.219 | 16.813 | 54.807 | 45.366 | 6.223 | 13.834 | 25.954 | 37.962 | 18.071 |
| 13 | 17.08 | 20.151 | 34.561 | 14.172 | 27.046 | 52.928 | 29.22 | 42.651 | 38.575 | 88.017 | 48.385 | 22.512 | 3.838 | 27.874 | 20.125 | 12.864 |
| 14 | 22.716 | 14.933 | 35.025 | 13.865 | 53.846 | 37.65 | 57.975 | 39.501 | 23.834 | 40.725 | 45.33 | 22.401 | 16.164 | 17.899 | 37.502 | 20.633 |
| 15 | 28.383 | 20.078 | 54.964 | 13.135 | 16.652 | 31.248 | 30.359 | 32.949 | 26.762 | 62.239 | 57.598 | 29.233 | 5.879 | 27.503 | 36.24 | 26.778 |
| 16 | 19.157 | 15.933 | 28.881 | 13.705 | 26.35 | 36.325 | 31.096 | 47.298 | 22.975 | 71.703 | 55.608 | 28.875 | 5.875 | 34.094 | 33.479 | 28.648 |
| 17 | 16.201 | 14.253 | 40.174 | 3.728 | 59.494 | 29.671 | 55.92 | 12.968 | 33.824 | 25.397 | 80.886 | 8.955 | 31.127 | 21.762 | 59.464 | 6.174 |
| 18 | 40.515 | 16.384 | 71.766 | 11.982 | 31.373 | 14.521 | 30.352 | 14.837 | 57.646 | 53.622 | 94.876 | 30.3 | 2.864 | 5.601 | 15.469 | 7.891 |
| 19 | 11.977 | 89.056 | 18.162 | 13.204 | 5.534 | 59.318 | 11.63 | 13.646 | 7.209 | 169.351 | 24.176 | 11.727 | 0.958 | 42.885 | 14.805 | 6.361 |
| 20 | 5.083 | 9.423 | 4.955 | 6.217 | 24.908 | 300.29 | 23.133 | 12.28 | 9.184 | 36.881 | 18.224 | 4.485 | 3.817 | 32.859 | 8.261 | 0.0 |
| 21 | 17.962 | 3.805 | 17.464 | 3.761 | 281.841 | 46.745 | 16.747 | 34.12 | 18.09 | 9.383 | 22.028 | 5.072 | 9.497 | 5.297 | 7.402 | 0.785 |
| 22 | 223.57 | 25.371 | 56.381 | 22.068 | 36.838 | 13.064 | 13.601 | 1.727 | 7.758 | 17.64 | 23.862 | 14.382 | 19.866 | 8.754 | 9.632 | 5.487 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.569 | -3.283 | 0.821 | -4.4 | -3.119 | -2.732 | 0.515 | -2.265 | -0.961 | -1.153 | 0.725 | -3.04 | -2.568 | -2.717 | 2.15 | -3.156 |
| 02 | -0.77 | -2.699 | -2.218 | -2.626 | -0.939 | -3.283 | -4.4 | -3.134 | 2.633 | -2.047 | -0.617 | -4.4 | -2.011 | -4.4 | -3.032 | -4.4 |
| 03 | -0.775 | 1.359 | 1.654 | 1.672 | -4.4 | -2.567 | -2.341 | -2.797 | -4.4 | -1.311 | -1.144 | -2.221 | -4.4 | -4.4 | -2.267 | -4.4 |
| 04 | -2.288 | -2.55 | -1.996 | -1.703 | -0.464 | -1.196 | 0.447 | 0.563 | -0.573 | 0.136 | -0.702 | 1.265 | -0.984 | -1.986 | 0.539 | 1.188 |
| 05 | -2.08 | -4.4 | 0.418 | -4.4 | -2.14 | -3.11 | 0.398 | -4.4 | -0.9 | -4.4 | 1.246 | -4.4 | -2.577 | -4.4 | 2.152 | -2.649 |
| 06 | -0.862 | -4.4 | -1.28 | -4.4 | -4.4 | -4.4 | -3.11 | -4.4 | 2.291 | -2.646 | 1.615 | -2.257 | -4.4 | -4.4 | -4.4 | -2.649 |
| 07 | -2.304 | 2.224 | -1.565 | -0.251 | -4.4 | -3.319 | -3.057 | -4.4 | -4.4 | 1.632 | -2.743 | -1.753 | -4.4 | -1.813 | -4.4 | -4.4 |
| 08 | -2.642 | -4.4 | -3.196 | -4.4 | 2.152 | -1.977 | 1.61 | -0.206 | -2.729 | -2.571 | -1.896 | -3.195 | -1.363 | -4.4 | -0.664 | -1.645 |
| 09 | -1.275 | -0.293 | 2.009 | -0.678 | -4.4 | -2.606 | -2.192 | -3.175 | -1.209 | -0.49 | 1.349 | -0.06 | -3.133 | -1.509 | -0.647 | -1.304 |
| 10 | -2.8 | -1.693 | -1.045 | -3.287 | -1.504 | -1.104 | -0.691 | -0.513 | 0.348 | 1.56 | 1.585 | -0.11 | -3.061 | -0.601 | -0.058 | -1.495 |
| 11 | -0.566 | -1.005 | -0.646 | -1.246 | -0.182 | 0.784 | 0.3 | 0.371 | 0.466 | 1.027 | 0.364 | -0.161 | -2.019 | -0.684 | -0.462 | -0.715 |
| 12 | -0.781 | -0.531 | 0.463 | -0.709 | 0.274 | 0.521 | 0.715 | -0.212 | -0.609 | 0.557 | 0.369 | -1.566 | -0.8 | -0.183 | 0.192 | -0.539 |
| 13 | -0.594 | -0.432 | 0.1 | -0.776 | -0.143 | 0.522 | -0.066 | 0.308 | 0.208 | 1.028 | 0.433 | -0.323 | -2.013 | -0.113 | -0.433 | -0.87 |
| 14 | -0.314 | -0.725 | 0.113 | -0.797 | 0.539 | 0.184 | 0.612 | 0.232 | -0.267 | 0.262 | 0.368 | -0.328 | -0.648 | -0.548 | 0.18 | -0.409 |
| 15 | -0.095 | -0.436 | 0.559 | -0.85 | -0.619 | -0.0 | -0.029 | 0.052 | -0.153 | 0.683 | 0.606 | -0.066 | -1.619 | -0.126 | 0.146 | -0.152 |
| 16 | -0.482 | -0.662 | -0.078 | -0.809 | -0.168 | 0.149 | -0.005 | 0.41 | -0.303 | 0.824 | 0.571 | -0.078 | -1.62 | 0.086 | 0.068 | -0.086 |
| 17 | -0.646 | -0.77 | 0.248 | -2.039 | 0.638 | -0.051 | 0.576 | -0.862 | 0.078 | -0.205 | 0.943 | -1.22 | -0.004 | -0.357 | 0.638 | -1.573 |
| 18 | 0.257 | -0.635 | 0.824 | -0.939 | 0.004 | -0.752 | -0.029 | -0.731 | 0.607 | 0.535 | 1.102 | -0.03 | -2.275 | -1.664 | -0.691 | -1.341 |
| 19 | -0.939 | 1.039 | -0.534 | -0.845 | -1.676 | 0.635 | -0.968 | -0.813 | -1.427 | 1.68 | -0.253 | -0.96 | -3.157 | 0.313 | -0.733 | -1.545 |
| 20 | -1.755 | -1.171 | -1.778 | -1.566 | -0.224 | 2.252 | -0.296 | -0.915 | -1.196 | 0.164 | -0.531 | -1.871 | -2.018 | 0.05 | -1.297 | -4.4 |
| 21 | -0.545 | -2.021 | -0.572 | -2.031 | 2.188 | 0.399 | -0.613 | 0.087 | -0.538 | -1.175 | -0.345 | -1.757 | -1.163 | -1.716 | -1.402 | -3.294 |
| 22 | 1.957 | -0.206 | 0.585 | -0.343 | 0.163 | -0.855 | -0.816 | -2.705 | -1.357 | -0.562 | -0.266 | -0.762 | -0.446 | -1.241 | -1.15 | -1.684 |