Model info
| Transcription factor | Pknox1 | ||||||||
| Model | PKNX1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 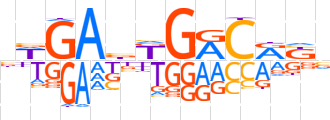 | ||||||||
| LOGO (reverse complement) | 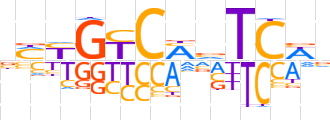 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bWGAbKGRCRKv | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.973 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 19 | ||||||||
| Aligned words | 479 | ||||||||
| TF family | TALE-type homeo domain factors {3.1.4} | ||||||||
| TF subfamily | PKNOX {3.1.4.5} | ||||||||
| MGI | MGI:1201409 | ||||||||
| EntrezGene | GeneID:18771 (SSTAR profile) | ||||||||
| UniProt ID | PKNX1_MOUSE | ||||||||
| UniProt AC | O70477 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Pknox1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 13.0 | 7.0 | 13.0 | 33.0 | 9.0 | 20.0 | 16.0 | 116.0 | 34.0 | 18.0 | 19.0 | 87.0 | 8.0 | 4.0 | 8.0 | 70.0 |
| 02 | 3.0 | 1.0 | 58.0 | 2.0 | 6.0 | 2.0 | 36.0 | 5.0 | 2.0 | 0.0 | 53.0 | 1.0 | 15.0 | 4.0 | 281.0 | 6.0 |
| 03 | 22.0 | 4.0 | 0.0 | 0.0 | 5.0 | 1.0 | 1.0 | 0.0 | 418.0 | 1.0 | 7.0 | 2.0 | 14.0 | 0.0 | 0.0 | 0.0 |
| 04 | 22.0 | 129.0 | 136.0 | 172.0 | 1.0 | 2.0 | 1.0 | 2.0 | 1.0 | 3.0 | 2.0 | 2.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 05 | 5.0 | 0.0 | 0.0 | 19.0 | 23.0 | 3.0 | 34.0 | 74.0 | 8.0 | 22.0 | 12.0 | 99.0 | 5.0 | 7.0 | 33.0 | 131.0 |
| 06 | 1.0 | 0.0 | 39.0 | 1.0 | 2.0 | 0.0 | 29.0 | 1.0 | 5.0 | 0.0 | 73.0 | 1.0 | 1.0 | 3.0 | 318.0 | 1.0 |
| 07 | 3.0 | 0.0 | 6.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 270.0 | 1.0 | 186.0 | 2.0 | 1.0 | 0.0 | 1.0 | 2.0 |
| 08 | 8.0 | 252.0 | 6.0 | 11.0 | 0.0 | 0.0 | 0.0 | 1.0 | 7.0 | 174.0 | 1.0 | 11.0 | 0.0 | 4.0 | 0.0 | 0.0 |
| 09 | 4.0 | 3.0 | 7.0 | 1.0 | 277.0 | 14.0 | 109.0 | 30.0 | 3.0 | 3.0 | 1.0 | 0.0 | 5.0 | 1.0 | 15.0 | 2.0 |
| 10 | 24.0 | 27.0 | 209.0 | 29.0 | 12.0 | 4.0 | 4.0 | 1.0 | 5.0 | 12.0 | 79.0 | 36.0 | 9.0 | 7.0 | 14.0 | 3.0 |
| 11 | 10.0 | 8.0 | 26.0 | 6.0 | 7.0 | 13.0 | 17.0 | 13.0 | 48.0 | 119.0 | 116.0 | 23.0 | 8.0 | 19.0 | 30.0 | 12.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.809 | -1.404 | -0.809 | 0.104 | -1.164 | -0.389 | -0.607 | 1.353 | 0.134 | -0.492 | -0.439 | 1.067 | -1.277 | -1.925 | -1.277 | 0.85 |
| 02 | -2.184 | -3.078 | 0.663 | -2.534 | -1.55 | -2.534 | 0.191 | -1.72 | -2.534 | -4.358 | 0.574 | -3.078 | -0.67 | -1.925 | 2.236 | -1.55 |
| 03 | -0.295 | -1.925 | -4.358 | -4.358 | -1.72 | -3.078 | -3.078 | -4.358 | 2.633 | -3.078 | -1.404 | -2.534 | -0.737 | -4.358 | -4.358 | -4.358 |
| 04 | -0.295 | 1.459 | 1.512 | 1.746 | -3.078 | -2.534 | -3.078 | -2.534 | -3.078 | -2.184 | -2.534 | -2.534 | -4.358 | -4.358 | -2.534 | -4.358 |
| 05 | -1.72 | -4.358 | -4.358 | -0.439 | -0.252 | -2.184 | 0.134 | 0.906 | -1.277 | -0.295 | -0.887 | 1.195 | -1.72 | -1.404 | 0.104 | 1.475 |
| 06 | -3.078 | -4.358 | 0.27 | -3.078 | -2.534 | -4.358 | -0.023 | -3.078 | -1.72 | -4.358 | 0.892 | -3.078 | -3.078 | -2.184 | 2.36 | -3.078 |
| 07 | -2.184 | -4.358 | -1.55 | -4.358 | -2.184 | -4.358 | -4.358 | -4.358 | 2.196 | -3.078 | 1.824 | -2.534 | -3.078 | -4.358 | -3.078 | -2.534 |
| 08 | -1.277 | 2.127 | -1.55 | -0.971 | -4.358 | -4.358 | -4.358 | -3.078 | -1.404 | 1.758 | -3.078 | -0.971 | -4.358 | -1.925 | -4.358 | -4.358 |
| 09 | -1.925 | -2.184 | -1.404 | -3.078 | 2.222 | -0.737 | 1.291 | 0.01 | -2.184 | -2.184 | -3.078 | -4.358 | -1.72 | -3.078 | -0.67 | -2.534 |
| 10 | -0.21 | -0.094 | 1.941 | -0.023 | -0.887 | -1.925 | -1.925 | -3.078 | -1.72 | -0.887 | 0.971 | 0.191 | -1.164 | -1.404 | -0.737 | -2.184 |
| 11 | -1.063 | -1.277 | -0.131 | -1.55 | -1.404 | -0.809 | -0.548 | -0.809 | 0.476 | 1.379 | 1.353 | -0.252 | -1.277 | -0.439 | 0.01 | -0.887 |