Model info
| Transcription factor | PLAGL1 (GeneCards) | ||||||||
| Model | PLAL1_HUMAN.H11MO.0.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 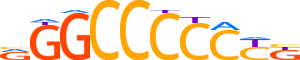 | ||||||||
| Data source | Integrative | ||||||||
| Model release | HOCOMOCOv9 | ||||||||
| Model length | 10 | ||||||||
| Quality | D | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vRGGGGGCCY | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 12 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | PLAG factors {2.3.3.25} | ||||||||
| HGNC | HGNC:9046 | ||||||||
| EntrezGene | GeneID:5325 (SSTAR profile) | ||||||||
| UniProt ID | PLAL1_HUMAN | ||||||||
| UniProt AC | Q9UM63 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | PLAGL1 expression | ||||||||
| ReMap ChIP-seq dataset list | PLAGL1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2.117 | 7.294 | 2.117 | 0.471 |
| 02 | 4.115 | 0.0 | 7.062 | 0.823 |
| 03 | 0.0 | 0.0 | 9.531 | 2.469 |
| 04 | 1.646 | 0.0 | 10.354 | 0.0 |
| 05 | 0.823 | 0.0 | 11.177 | 0.0 |
| 06 | 0.0 | 0.0 | 12.0 | 0.0 |
| 07 | 0.0 | 0.0 | 12.0 | 0.0 |
| 08 | 0.0 | 11.177 | 0.0 | 0.823 |
| 09 | 0.823 | 10.354 | 0.0 | 0.823 |
| 10 | 0.823 | 7.062 | 1.646 | 2.469 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.279 | 0.782 | -0.279 | -1.198 |
| 02 | 0.268 | -1.763 | 0.752 | -0.919 |
| 03 | -1.763 | -1.763 | 1.031 | -0.159 |
| 04 | -0.468 | -1.763 | 1.109 | -1.763 |
| 05 | -0.919 | -1.763 | 1.181 | -1.763 |
| 06 | -1.763 | -1.763 | 1.249 | -1.763 |
| 07 | -1.763 | -1.763 | 1.249 | -1.763 |
| 08 | -1.763 | 1.181 | -1.763 | -0.919 |
| 09 | -0.919 | 1.109 | -1.763 | -0.919 |
| 10 | -0.919 | 0.752 | -0.468 | -0.159 |