Model info
| Transcription factor | Pou3f1 | ||||||||
| Model | PO3F1_MOUSE.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 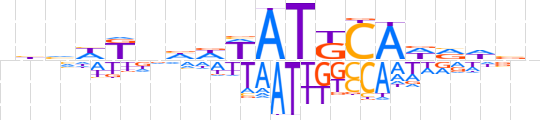 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbdbWTGMATWhnnMWdvn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.849 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 10 | ||||||||
| Aligned words | 504 | ||||||||
| TF family | POU domain factors {3.1.10} | ||||||||
| TF subfamily | POU3 (Oct-6-like factors) {3.1.10.3} | ||||||||
| MGI | MGI:101896 | ||||||||
| EntrezGene | GeneID:18991 (SSTAR profile) | ||||||||
| UniProt ID | PO3F1_MOUSE | ||||||||
| UniProt AC | P21952 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Pou3f1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 21.0 | 34.0 | 41.0 | 10.0 | 9.0 | 52.0 | 7.0 | 31.0 | 9.0 | 59.0 | 26.0 | 22.0 | 19.0 | 76.0 | 57.0 | 26.0 |
| 02 | 36.0 | 4.0 | 6.0 | 12.0 | 99.0 | 10.0 | 5.0 | 107.0 | 61.0 | 19.0 | 21.0 | 30.0 | 30.0 | 5.0 | 15.0 | 39.0 |
| 03 | 16.0 | 39.0 | 16.0 | 155.0 | 3.0 | 20.0 | 1.0 | 14.0 | 6.0 | 25.0 | 9.0 | 7.0 | 7.0 | 129.0 | 17.0 | 35.0 |
| 04 | 22.0 | 4.0 | 3.0 | 3.0 | 167.0 | 20.0 | 1.0 | 25.0 | 20.0 | 18.0 | 2.0 | 3.0 | 25.0 | 23.0 | 8.0 | 155.0 |
| 05 | 6.0 | 2.0 | 2.0 | 224.0 | 3.0 | 1.0 | 0.0 | 61.0 | 2.0 | 2.0 | 1.0 | 9.0 | 47.0 | 1.0 | 5.0 | 133.0 |
| 06 | 6.0 | 8.0 | 44.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 1.0 | 2.0 | 5.0 | 0.0 | 24.0 | 18.0 | 384.0 | 1.0 |
| 07 | 14.0 | 16.0 | 0.0 | 1.0 | 3.0 | 25.0 | 0.0 | 0.0 | 192.0 | 244.0 | 2.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 08 | 206.0 | 0.0 | 2.0 | 2.0 | 284.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 09 | 13.0 | 2.0 | 3.0 | 475.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 |
| 10 | 13.0 | 1.0 | 1.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 346.0 | 24.0 | 41.0 | 67.0 |
| 11 | 98.0 | 39.0 | 14.0 | 212.0 | 11.0 | 6.0 | 0.0 | 8.0 | 10.0 | 19.0 | 2.0 | 12.0 | 26.0 | 2.0 | 11.0 | 29.0 |
| 12 | 27.0 | 27.0 | 18.0 | 73.0 | 27.0 | 11.0 | 2.0 | 26.0 | 4.0 | 6.0 | 3.0 | 14.0 | 23.0 | 19.0 | 58.0 | 161.0 |
| 13 | 30.0 | 24.0 | 15.0 | 12.0 | 43.0 | 7.0 | 0.0 | 13.0 | 26.0 | 27.0 | 8.0 | 20.0 | 57.0 | 98.0 | 42.0 | 77.0 |
| 14 | 117.0 | 21.0 | 8.0 | 10.0 | 136.0 | 11.0 | 0.0 | 9.0 | 36.0 | 21.0 | 4.0 | 4.0 | 63.0 | 40.0 | 4.0 | 15.0 |
| 15 | 97.0 | 17.0 | 36.0 | 202.0 | 69.0 | 6.0 | 2.0 | 16.0 | 8.0 | 3.0 | 3.0 | 2.0 | 14.0 | 3.0 | 3.0 | 18.0 |
| 16 | 41.0 | 12.0 | 54.0 | 81.0 | 11.0 | 7.0 | 5.0 | 6.0 | 16.0 | 7.0 | 5.0 | 16.0 | 36.0 | 22.0 | 125.0 | 55.0 |
| 17 | 30.0 | 14.0 | 41.0 | 19.0 | 25.0 | 5.0 | 11.0 | 7.0 | 96.0 | 33.0 | 43.0 | 17.0 | 43.0 | 29.0 | 63.0 | 23.0 |
| 18 | 48.0 | 36.0 | 86.0 | 24.0 | 45.0 | 15.0 | 9.0 | 12.0 | 51.0 | 51.0 | 34.0 | 22.0 | 18.0 | 14.0 | 14.0 | 20.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.39 | 0.085 | 0.271 | -1.112 | -1.213 | 0.506 | -1.452 | -0.006 | -1.213 | 0.632 | -0.179 | -0.344 | -0.488 | 0.883 | 0.597 | -0.179 |
| 02 | 0.142 | -1.973 | -1.598 | -0.936 | 1.147 | -1.112 | -1.768 | 1.224 | 0.665 | -0.488 | -0.39 | -0.038 | -0.038 | -1.768 | -0.719 | 0.221 |
| 03 | -0.656 | 0.221 | -0.656 | 1.594 | -2.232 | -0.437 | -3.124 | -0.786 | -1.598 | -0.218 | -1.213 | -1.452 | -1.452 | 1.41 | -0.597 | 0.114 |
| 04 | -0.344 | -1.973 | -2.232 | -2.232 | 1.668 | -0.437 | -3.124 | -0.218 | -0.437 | -0.541 | -2.582 | -2.232 | -0.218 | -0.3 | -1.326 | 1.594 |
| 05 | -1.598 | -2.582 | -2.582 | 1.961 | -2.232 | -3.124 | -4.398 | 0.665 | -2.582 | -2.582 | -3.124 | -1.213 | 0.406 | -3.124 | -1.768 | 1.441 |
| 06 | -1.598 | -1.326 | 0.341 | -4.398 | -4.398 | -4.398 | -1.598 | -4.398 | -3.124 | -2.582 | -1.768 | -4.398 | -0.258 | -0.541 | 2.499 | -3.124 |
| 07 | -0.786 | -0.656 | -4.398 | -3.124 | -2.232 | -0.218 | -4.398 | -4.398 | 1.807 | 2.046 | -2.582 | -3.124 | -3.124 | -4.398 | -4.398 | -4.398 |
| 08 | 1.877 | -4.398 | -2.582 | -2.582 | 2.198 | -3.124 | -4.398 | -4.398 | -3.124 | -4.398 | -4.398 | -3.124 | -2.582 | -4.398 | -4.398 | -4.398 |
| 09 | -0.858 | -2.582 | -2.232 | 2.712 | -4.398 | -3.124 | -4.398 | -4.398 | -2.582 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -2.232 |
| 10 | -0.858 | -3.124 | -3.124 | -4.398 | -2.232 | -4.398 | -4.398 | -4.398 | -3.124 | -4.398 | -3.124 | -3.124 | 2.395 | -0.258 | 0.271 | 0.758 |
| 11 | 1.137 | 0.221 | -0.786 | 1.906 | -1.02 | -1.598 | -4.398 | -1.326 | -1.112 | -0.488 | -2.582 | -0.936 | -0.179 | -2.582 | -1.02 | -0.072 |
| 12 | -0.142 | -0.142 | -0.541 | 0.843 | -0.142 | -1.02 | -2.582 | -0.179 | -1.973 | -1.598 | -2.232 | -0.786 | -0.3 | -0.488 | 0.615 | 1.631 |
| 13 | -0.038 | -0.258 | -0.719 | -0.936 | 0.318 | -1.452 | -4.398 | -0.858 | -0.179 | -0.142 | -1.326 | -0.437 | 0.597 | 1.137 | 0.294 | 0.896 |
| 14 | 1.313 | -0.39 | -1.326 | -1.112 | 1.463 | -1.02 | -4.398 | -1.213 | 0.142 | -0.39 | -1.973 | -1.973 | 0.697 | 0.246 | -1.973 | -0.719 |
| 15 | 1.126 | -0.597 | 0.142 | 1.858 | 0.787 | -1.598 | -2.582 | -0.656 | -1.326 | -2.232 | -2.232 | -2.582 | -0.786 | -2.232 | -2.232 | -0.541 |
| 16 | 0.271 | -0.936 | 0.544 | 0.947 | -1.02 | -1.452 | -1.768 | -1.598 | -0.656 | -1.452 | -1.768 | -0.656 | 0.142 | -0.344 | 1.379 | 0.562 |
| 17 | -0.038 | -0.786 | 0.271 | -0.488 | -0.218 | -1.768 | -1.02 | -1.452 | 1.116 | 0.056 | 0.318 | -0.597 | 0.318 | -0.072 | 0.697 | -0.3 |
| 18 | 0.427 | 0.142 | 1.006 | -0.258 | 0.363 | -0.719 | -1.213 | -0.936 | 0.487 | 0.487 | 0.085 | -0.344 | -0.541 | -0.786 | -0.786 | -0.437 |