Model info
| Transcription factor | Pou3f2 | ||||||||
| Model | PO3F2_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 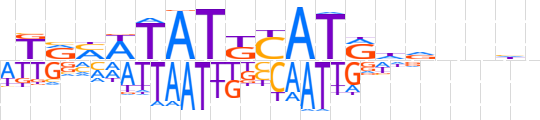 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bdnnbdYATGMATAWKYMW | ||||||||
| Best auROC (human) | 0.787 | ||||||||
| Best auROC (mouse) | 0.956 | ||||||||
| Peak sets in benchmark (human) | 12 | ||||||||
| Peak sets in benchmark (mouse) | 22 | ||||||||
| Aligned words | 385 | ||||||||
| TF family | POU domain factors {3.1.10} | ||||||||
| TF subfamily | POU3 (Oct-6-like factors) {3.1.10.3} | ||||||||
| MGI | MGI:101895 | ||||||||
| EntrezGene | GeneID:18992 (SSTAR profile) | ||||||||
| UniProt ID | PO3F2_MOUSE | ||||||||
| UniProt AC | P31360 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Pou3f2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 30.0 | 7.0 | 13.0 | 12.0 | 44.0 | 15.0 | 6.0 | 33.0 | 26.0 | 9.0 | 19.0 | 19.0 | 54.0 | 5.0 | 56.0 | 33.0 |
| 02 | 43.0 | 34.0 | 20.0 | 57.0 | 15.0 | 11.0 | 1.0 | 9.0 | 26.0 | 20.0 | 15.0 | 33.0 | 25.0 | 19.0 | 23.0 | 30.0 |
| 03 | 40.0 | 17.0 | 28.0 | 24.0 | 39.0 | 22.0 | 4.0 | 19.0 | 13.0 | 23.0 | 10.0 | 13.0 | 44.0 | 20.0 | 35.0 | 30.0 |
| 04 | 9.0 | 57.0 | 13.0 | 57.0 | 11.0 | 30.0 | 2.0 | 39.0 | 7.0 | 38.0 | 10.0 | 22.0 | 8.0 | 38.0 | 16.0 | 24.0 |
| 05 | 15.0 | 8.0 | 4.0 | 8.0 | 61.0 | 7.0 | 5.0 | 90.0 | 11.0 | 9.0 | 5.0 | 16.0 | 59.0 | 19.0 | 31.0 | 33.0 |
| 06 | 12.0 | 90.0 | 3.0 | 41.0 | 7.0 | 28.0 | 0.0 | 8.0 | 3.0 | 26.0 | 1.0 | 15.0 | 11.0 | 115.0 | 2.0 | 19.0 |
| 07 | 32.0 | 1.0 | 0.0 | 0.0 | 246.0 | 3.0 | 0.0 | 10.0 | 6.0 | 0.0 | 0.0 | 0.0 | 71.0 | 3.0 | 0.0 | 9.0 |
| 08 | 1.0 | 3.0 | 3.0 | 348.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 2.0 | 15.0 |
| 09 | 1.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 2.0 | 0.0 | 1.0 | 2.0 | 2.0 | 0.0 | 85.0 | 6.0 | 275.0 | 4.0 |
| 10 | 43.0 | 45.0 | 1.0 | 1.0 | 2.0 | 5.0 | 0.0 | 1.0 | 134.0 | 134.0 | 2.0 | 9.0 | 2.0 | 2.0 | 0.0 | 0.0 |
| 11 | 179.0 | 0.0 | 2.0 | 0.0 | 178.0 | 4.0 | 0.0 | 4.0 | 1.0 | 1.0 | 0.0 | 1.0 | 8.0 | 1.0 | 0.0 | 2.0 |
| 12 | 5.0 | 7.0 | 1.0 | 353.0 | 0.0 | 1.0 | 0.0 | 5.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 5.0 |
| 13 | 4.0 | 0.0 | 0.0 | 2.0 | 8.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 320.0 | 2.0 | 2.0 | 40.0 |
| 14 | 58.0 | 17.0 | 5.0 | 253.0 | 2.0 | 0.0 | 0.0 | 2.0 | 0.0 | 1.0 | 0.0 | 1.0 | 25.0 | 2.0 | 1.0 | 14.0 |
| 15 | 12.0 | 1.0 | 2.0 | 70.0 | 8.0 | 1.0 | 1.0 | 10.0 | 0.0 | 0.0 | 3.0 | 3.0 | 14.0 | 5.0 | 130.0 | 121.0 |
| 16 | 5.0 | 18.0 | 1.0 | 10.0 | 1.0 | 6.0 | 0.0 | 0.0 | 2.0 | 108.0 | 9.0 | 17.0 | 9.0 | 113.0 | 48.0 | 34.0 |
| 17 | 9.0 | 7.0 | 1.0 | 0.0 | 234.0 | 3.0 | 1.0 | 7.0 | 10.0 | 41.0 | 7.0 | 0.0 | 29.0 | 19.0 | 6.0 | 7.0 |
| 18 | 43.0 | 7.0 | 15.0 | 217.0 | 62.0 | 3.0 | 1.0 | 4.0 | 7.0 | 6.0 | 0.0 | 2.0 | 6.0 | 2.0 | 1.0 | 5.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.228 | -1.188 | -0.593 | -0.67 | 0.607 | -0.453 | -1.334 | 0.322 | 0.087 | -0.948 | -0.222 | -0.222 | 0.81 | -1.505 | 0.846 | 0.322 |
| 02 | 0.584 | 0.352 | -0.172 | 0.864 | -0.453 | -0.755 | -2.87 | -0.948 | 0.087 | -0.172 | -0.453 | 0.322 | 0.048 | -0.222 | -0.034 | 0.228 |
| 03 | 0.512 | -0.331 | 0.16 | 0.008 | 0.487 | -0.078 | -1.711 | -0.222 | -0.593 | -0.034 | -0.847 | -0.593 | 0.607 | -0.172 | 0.38 | 0.228 |
| 04 | -0.948 | 0.864 | -0.593 | 0.864 | -0.755 | 0.228 | -2.322 | 0.487 | -1.188 | 0.462 | -0.847 | -0.078 | -1.061 | 0.462 | -0.39 | 0.008 |
| 05 | -0.453 | -1.061 | -1.711 | -1.061 | 0.931 | -1.188 | -1.505 | 1.318 | -0.755 | -0.948 | -1.505 | -0.39 | 0.898 | -0.222 | 0.26 | 0.322 |
| 06 | -0.67 | 1.318 | -1.97 | 0.537 | -1.188 | 0.16 | -4.176 | -1.061 | -1.97 | 0.087 | -2.87 | -0.453 | -0.755 | 1.562 | -2.322 | -0.222 |
| 07 | 0.292 | -2.87 | -4.176 | -4.176 | 2.321 | -1.97 | -4.176 | -0.847 | -1.334 | -4.176 | -4.176 | -4.176 | 1.082 | -1.97 | -4.176 | -0.948 |
| 08 | -2.87 | -1.97 | -1.97 | 2.668 | -4.176 | -4.176 | -4.176 | -1.188 | -4.176 | -4.176 | -4.176 | -4.176 | -4.176 | -2.322 | -2.322 | -0.453 |
| 09 | -2.87 | -4.176 | -4.176 | -4.176 | -1.97 | -4.176 | -2.322 | -4.176 | -2.87 | -2.322 | -2.322 | -4.176 | 1.261 | -1.334 | 2.432 | -1.711 |
| 10 | 0.584 | 0.629 | -2.87 | -2.87 | -2.322 | -1.505 | -4.176 | -2.87 | 1.715 | 1.715 | -2.322 | -0.948 | -2.322 | -2.322 | -4.176 | -4.176 |
| 11 | 2.004 | -4.176 | -2.322 | -4.176 | 1.998 | -1.711 | -4.176 | -1.711 | -2.87 | -2.87 | -4.176 | -2.87 | -1.061 | -2.87 | -4.176 | -2.322 |
| 12 | -1.505 | -1.188 | -2.87 | 2.682 | -4.176 | -2.87 | -4.176 | -1.505 | -2.87 | -4.176 | -4.176 | -2.87 | -4.176 | -2.87 | -2.87 | -1.505 |
| 13 | -1.711 | -4.176 | -4.176 | -2.322 | -1.061 | -2.87 | -4.176 | -4.176 | -2.87 | -2.87 | -4.176 | -4.176 | 2.584 | -2.322 | -2.322 | 0.512 |
| 14 | 0.881 | -0.331 | -1.505 | 2.349 | -2.322 | -4.176 | -4.176 | -2.322 | -4.176 | -2.87 | -4.176 | -2.87 | 0.048 | -2.322 | -2.87 | -0.52 |
| 15 | -0.67 | -2.87 | -2.322 | 1.068 | -1.061 | -2.87 | -2.87 | -0.847 | -4.176 | -4.176 | -1.97 | -1.97 | -0.52 | -1.505 | 1.685 | 1.613 |
| 16 | -1.505 | -0.275 | -2.87 | -0.847 | -2.87 | -1.334 | -4.176 | -4.176 | -2.322 | 1.5 | -0.948 | -0.331 | -0.948 | 1.545 | 0.693 | 0.352 |
| 17 | -0.948 | -1.188 | -2.87 | -4.176 | 2.271 | -1.97 | -2.87 | -1.188 | -0.847 | 0.537 | -1.188 | -4.176 | 0.194 | -0.222 | -1.334 | -1.188 |
| 18 | 0.584 | -1.188 | -0.453 | 2.196 | 0.947 | -1.97 | -2.87 | -1.711 | -1.188 | -1.334 | -4.176 | -2.322 | -1.334 | -2.322 | -2.87 | -1.505 |