Model info
| Transcription factor | PPARA (GeneCards) | ||||||||
| Model | PPARA_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 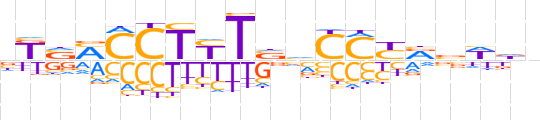 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nhWbYRKGbbARAGGKbMn | ||||||||
| Best auROC (human) | 0.59 | ||||||||
| Best auROC (mouse) | 0.965 | ||||||||
| Peak sets in benchmark (human) | 5 | ||||||||
| Peak sets in benchmark (mouse) | 20 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | PPAR (NR1C) {2.1.2.5} | ||||||||
| HGNC | HGNC:9232 | ||||||||
| EntrezGene | GeneID:5465 (SSTAR profile) | ||||||||
| UniProt ID | PPARA_HUMAN | ||||||||
| UniProt AC | Q07869 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | PPARA expression | ||||||||
| ReMap ChIP-seq dataset list | PPARA datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 92.0 | 21.0 | 25.0 | 13.0 | 77.0 | 21.0 | 2.0 | 21.0 | 46.0 | 24.0 | 23.0 | 17.0 | 35.0 | 34.0 | 21.0 | 28.0 |
| 02 | 157.0 | 11.0 | 26.0 | 56.0 | 66.0 | 4.0 | 3.0 | 27.0 | 44.0 | 5.0 | 10.0 | 12.0 | 21.0 | 11.0 | 8.0 | 39.0 |
| 03 | 29.0 | 107.0 | 120.0 | 32.0 | 10.0 | 7.0 | 3.0 | 11.0 | 8.0 | 18.0 | 11.0 | 10.0 | 13.0 | 50.0 | 52.0 | 19.0 |
| 04 | 6.0 | 8.0 | 13.0 | 33.0 | 21.0 | 32.0 | 6.0 | 123.0 | 12.0 | 30.0 | 25.0 | 119.0 | 9.0 | 18.0 | 19.0 | 26.0 |
| 05 | 24.0 | 0.0 | 19.0 | 5.0 | 70.0 | 4.0 | 7.0 | 7.0 | 33.0 | 4.0 | 23.0 | 3.0 | 95.0 | 6.0 | 191.0 | 9.0 |
| 06 | 32.0 | 0.0 | 180.0 | 10.0 | 2.0 | 0.0 | 6.0 | 6.0 | 16.0 | 3.0 | 177.0 | 44.0 | 2.0 | 0.0 | 20.0 | 2.0 |
| 07 | 6.0 | 3.0 | 41.0 | 2.0 | 1.0 | 1.0 | 0.0 | 1.0 | 55.0 | 7.0 | 310.0 | 11.0 | 5.0 | 4.0 | 50.0 | 3.0 |
| 08 | 6.0 | 10.0 | 41.0 | 10.0 | 4.0 | 6.0 | 0.0 | 5.0 | 37.0 | 92.0 | 141.0 | 131.0 | 5.0 | 7.0 | 1.0 | 4.0 |
| 09 | 4.0 | 29.0 | 14.0 | 5.0 | 14.0 | 70.0 | 8.0 | 23.0 | 16.0 | 108.0 | 37.0 | 22.0 | 16.0 | 73.0 | 44.0 | 17.0 |
| 10 | 41.0 | 1.0 | 8.0 | 0.0 | 271.0 | 6.0 | 2.0 | 1.0 | 93.0 | 0.0 | 9.0 | 1.0 | 59.0 | 6.0 | 2.0 | 0.0 |
| 11 | 288.0 | 14.0 | 136.0 | 26.0 | 10.0 | 1.0 | 1.0 | 1.0 | 14.0 | 1.0 | 5.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| 12 | 258.0 | 0.0 | 53.0 | 2.0 | 14.0 | 0.0 | 2.0 | 0.0 | 113.0 | 0.0 | 30.0 | 0.0 | 15.0 | 2.0 | 11.0 | 0.0 |
| 13 | 50.0 | 1.0 | 342.0 | 7.0 | 0.0 | 0.0 | 1.0 | 1.0 | 22.0 | 0.0 | 71.0 | 3.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 14 | 2.0 | 0.0 | 70.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 15.0 | 2.0 | 312.0 | 87.0 | 0.0 | 0.0 | 11.0 | 0.0 |
| 15 | 1.0 | 0.0 | 10.0 | 6.0 | 0.0 | 0.0 | 0.0 | 2.0 | 18.0 | 42.0 | 79.0 | 255.0 | 1.0 | 4.0 | 19.0 | 63.0 |
| 16 | 2.0 | 14.0 | 4.0 | 0.0 | 10.0 | 23.0 | 2.0 | 11.0 | 13.0 | 62.0 | 19.0 | 14.0 | 20.0 | 193.0 | 60.0 | 53.0 |
| 17 | 29.0 | 8.0 | 5.0 | 3.0 | 230.0 | 28.0 | 12.0 | 22.0 | 67.0 | 9.0 | 6.0 | 3.0 | 39.0 | 12.0 | 22.0 | 5.0 |
| 18 | 77.0 | 119.0 | 112.0 | 57.0 | 20.0 | 13.0 | 4.0 | 20.0 | 8.0 | 9.0 | 9.0 | 19.0 | 7.0 | 10.0 | 7.0 | 9.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.072 | -0.392 | -0.22 | -0.86 | 0.894 | -0.392 | -2.584 | -0.392 | 0.383 | -0.26 | -0.302 | -0.599 | 0.112 | 0.083 | -0.392 | -0.108 |
| 02 | 1.604 | -1.022 | -0.181 | 0.578 | 0.741 | -1.975 | -2.234 | -0.144 | 0.339 | -1.77 | -1.114 | -0.938 | -0.392 | -1.022 | -1.328 | 0.219 |
| 03 | -0.074 | 1.222 | 1.336 | 0.023 | -1.114 | -1.454 | -2.234 | -1.022 | -1.328 | -0.543 | -1.022 | -1.114 | -0.86 | 0.465 | 0.504 | -0.49 |
| 04 | -1.6 | -1.328 | -0.86 | 0.054 | -0.392 | 0.023 | -1.6 | 1.361 | -0.938 | -0.04 | -0.22 | 1.328 | -1.215 | -0.543 | -0.49 | -0.181 |
| 05 | -0.26 | -4.4 | -0.49 | -1.77 | 0.8 | -1.975 | -1.454 | -1.454 | 0.054 | -1.975 | -0.302 | -2.234 | 1.104 | -1.6 | 1.8 | -1.215 |
| 06 | 0.023 | -4.4 | 1.741 | -1.114 | -2.584 | -4.4 | -1.6 | -1.6 | -0.658 | -2.234 | 1.724 | 0.339 | -2.584 | -4.4 | -0.439 | -2.584 |
| 07 | -1.6 | -2.234 | 0.269 | -2.584 | -3.126 | -3.126 | -4.4 | -3.126 | 0.56 | -1.454 | 2.283 | -1.022 | -1.77 | -1.975 | 0.465 | -2.234 |
| 08 | -1.6 | -1.114 | 0.269 | -1.114 | -1.975 | -1.6 | -4.4 | -1.77 | 0.167 | 1.072 | 1.497 | 1.424 | -1.77 | -1.454 | -3.126 | -1.975 |
| 09 | -1.975 | -0.074 | -0.788 | -1.77 | -0.788 | 0.8 | -1.328 | -0.302 | -0.658 | 1.231 | 0.167 | -0.346 | -0.658 | 0.841 | 0.339 | -0.599 |
| 10 | 0.269 | -3.126 | -1.328 | -4.4 | 2.149 | -1.6 | -2.584 | -3.126 | 1.082 | -4.4 | -1.215 | -3.126 | 0.63 | -1.6 | -2.584 | -4.4 |
| 11 | 2.21 | -0.788 | 1.461 | -0.181 | -1.114 | -3.126 | -3.126 | -3.126 | -0.788 | -3.126 | -1.77 | -3.126 | -3.126 | -4.4 | -3.126 | -4.4 |
| 12 | 2.1 | -4.4 | 0.523 | -2.584 | -0.788 | -4.4 | -2.584 | -4.4 | 1.276 | -4.4 | -0.04 | -4.4 | -0.721 | -2.584 | -1.022 | -4.4 |
| 13 | 0.465 | -3.126 | 2.382 | -1.454 | -4.4 | -4.4 | -3.126 | -3.126 | -0.346 | -4.4 | 0.814 | -2.234 | -4.4 | -4.4 | -2.584 | -4.4 |
| 14 | -2.584 | -4.4 | 0.8 | -4.4 | -4.4 | -4.4 | -3.126 | -4.4 | -0.721 | -2.584 | 2.29 | 1.016 | -4.4 | -4.4 | -1.022 | -4.4 |
| 15 | -3.126 | -4.4 | -1.114 | -1.6 | -4.4 | -4.4 | -4.4 | -2.584 | -0.543 | 0.293 | 0.92 | 2.088 | -3.126 | -1.975 | -0.49 | 0.695 |
| 16 | -2.584 | -0.788 | -1.975 | -4.4 | -1.114 | -0.302 | -2.584 | -1.022 | -0.86 | 0.679 | -0.49 | -0.788 | -0.439 | 1.81 | 0.646 | 0.523 |
| 17 | -0.074 | -1.328 | -1.77 | -2.234 | 1.985 | -0.108 | -0.938 | -0.346 | 0.756 | -1.215 | -1.6 | -2.234 | 0.219 | -0.938 | -0.346 | -1.77 |
| 18 | 0.894 | 1.328 | 1.268 | 0.595 | -0.439 | -0.86 | -1.975 | -0.439 | -1.328 | -1.215 | -1.215 | -0.49 | -1.454 | -1.114 | -1.454 | -1.215 |