Model info
| Transcription factor | PPARG (GeneCards) | ||||||||
| Model | PPARG_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 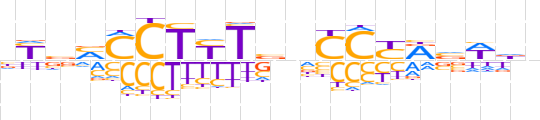 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nhWbYRGRnbARAGKKbRn | ||||||||
| Best auROC (human) | 0.963 | ||||||||
| Best auROC (mouse) | 0.963 | ||||||||
| Peak sets in benchmark (human) | 53 | ||||||||
| Peak sets in benchmark (mouse) | 114 | ||||||||
| Aligned words | 486 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | PPAR (NR1C) {2.1.2.5} | ||||||||
| HGNC | HGNC:9236 | ||||||||
| EntrezGene | GeneID:5468 (SSTAR profile) | ||||||||
| UniProt ID | PPARG_HUMAN | ||||||||
| UniProt AC | P37231 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | PPARG expression | ||||||||
| ReMap ChIP-seq dataset list | PPARG datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 72.0 | 14.0 | 18.0 | 16.0 | 80.0 | 22.0 | 3.0 | 22.0 | 53.0 | 29.0 | 20.0 | 14.0 | 29.0 | 36.0 | 12.0 | 44.0 |
| 02 | 144.0 | 7.0 | 13.0 | 70.0 | 48.0 | 1.0 | 6.0 | 46.0 | 27.0 | 0.0 | 7.0 | 19.0 | 27.0 | 2.0 | 6.0 | 61.0 |
| 03 | 23.0 | 99.0 | 92.0 | 32.0 | 3.0 | 1.0 | 1.0 | 5.0 | 2.0 | 14.0 | 15.0 | 1.0 | 10.0 | 80.0 | 86.0 | 20.0 |
| 04 | 7.0 | 3.0 | 3.0 | 25.0 | 20.0 | 22.0 | 6.0 | 146.0 | 14.0 | 20.0 | 26.0 | 134.0 | 8.0 | 5.0 | 14.0 | 31.0 |
| 05 | 21.0 | 3.0 | 23.0 | 2.0 | 41.0 | 3.0 | 2.0 | 4.0 | 29.0 | 3.0 | 15.0 | 2.0 | 105.0 | 2.0 | 213.0 | 16.0 |
| 06 | 16.0 | 3.0 | 164.0 | 13.0 | 0.0 | 0.0 | 11.0 | 0.0 | 10.0 | 2.0 | 205.0 | 36.0 | 1.0 | 0.0 | 21.0 | 2.0 |
| 07 | 6.0 | 0.0 | 19.0 | 2.0 | 1.0 | 0.0 | 4.0 | 0.0 | 78.0 | 8.0 | 307.0 | 8.0 | 7.0 | 1.0 | 38.0 | 5.0 |
| 08 | 29.0 | 13.0 | 44.0 | 6.0 | 2.0 | 0.0 | 3.0 | 4.0 | 50.0 | 68.0 | 124.0 | 126.0 | 7.0 | 3.0 | 4.0 | 1.0 |
| 09 | 5.0 | 41.0 | 26.0 | 16.0 | 18.0 | 40.0 | 10.0 | 16.0 | 25.0 | 83.0 | 53.0 | 14.0 | 7.0 | 42.0 | 68.0 | 20.0 |
| 10 | 42.0 | 3.0 | 7.0 | 3.0 | 189.0 | 6.0 | 3.0 | 8.0 | 138.0 | 3.0 | 13.0 | 3.0 | 54.0 | 2.0 | 8.0 | 2.0 |
| 11 | 273.0 | 10.0 | 116.0 | 24.0 | 8.0 | 0.0 | 3.0 | 3.0 | 21.0 | 2.0 | 7.0 | 1.0 | 6.0 | 2.0 | 6.0 | 2.0 |
| 12 | 263.0 | 0.0 | 43.0 | 2.0 | 12.0 | 0.0 | 2.0 | 0.0 | 101.0 | 1.0 | 30.0 | 0.0 | 16.0 | 3.0 | 10.0 | 1.0 |
| 13 | 38.0 | 0.0 | 349.0 | 5.0 | 0.0 | 0.0 | 3.0 | 1.0 | 15.0 | 1.0 | 68.0 | 1.0 | 1.0 | 0.0 | 2.0 | 0.0 |
| 14 | 2.0 | 1.0 | 48.0 | 3.0 | 0.0 | 0.0 | 1.0 | 0.0 | 31.0 | 8.0 | 323.0 | 60.0 | 0.0 | 0.0 | 7.0 | 0.0 |
| 15 | 3.0 | 1.0 | 23.0 | 6.0 | 2.0 | 1.0 | 3.0 | 3.0 | 17.0 | 43.0 | 134.0 | 185.0 | 2.0 | 1.0 | 12.0 | 48.0 |
| 16 | 2.0 | 11.0 | 8.0 | 3.0 | 13.0 | 17.0 | 2.0 | 14.0 | 38.0 | 66.0 | 36.0 | 32.0 | 23.0 | 130.0 | 61.0 | 28.0 |
| 17 | 49.0 | 6.0 | 13.0 | 8.0 | 168.0 | 11.0 | 21.0 | 24.0 | 74.0 | 6.0 | 15.0 | 12.0 | 43.0 | 8.0 | 19.0 | 7.0 |
| 18 | 97.0 | 76.0 | 108.0 | 53.0 | 6.0 | 9.0 | 3.0 | 13.0 | 15.0 | 22.0 | 17.0 | 14.0 | 9.0 | 9.0 | 12.0 | 21.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.86 | -0.756 | -0.511 | -0.626 | 0.965 | -0.314 | -2.202 | -0.314 | 0.555 | -0.042 | -0.407 | -0.756 | -0.042 | 0.172 | -0.906 | 0.371 |
| 02 | 1.55 | -1.423 | -0.828 | 0.832 | 0.457 | -3.095 | -1.568 | 0.415 | -0.112 | -4.373 | -1.423 | -0.458 | -0.112 | -2.552 | -1.568 | 0.695 |
| 03 | -0.27 | 1.177 | 1.104 | 0.056 | -2.202 | -3.095 | -3.095 | -1.738 | -2.552 | -0.756 | -0.689 | -3.095 | -1.082 | 0.965 | 1.037 | -0.407 |
| 04 | -1.423 | -2.202 | -2.202 | -0.188 | -0.407 | -0.314 | -1.568 | 1.564 | -0.756 | -0.407 | -0.149 | 1.479 | -1.296 | -1.738 | -0.756 | 0.024 |
| 05 | -0.359 | -2.202 | -0.27 | -2.552 | 0.301 | -2.202 | -2.552 | -1.944 | -0.042 | -2.202 | -0.689 | -2.552 | 1.235 | -2.552 | 1.941 | -0.626 |
| 06 | -0.626 | -2.202 | 1.68 | -0.828 | -4.373 | -4.373 | -0.99 | -4.373 | -1.082 | -2.552 | 1.903 | 0.172 | -3.095 | -4.373 | -0.359 | -2.552 |
| 07 | -1.568 | -4.373 | -0.458 | -2.552 | -3.095 | -4.373 | -1.944 | -4.373 | 0.939 | -1.296 | 2.306 | -1.296 | -1.423 | -3.095 | 0.226 | -1.738 |
| 08 | -0.042 | -0.828 | 0.371 | -1.568 | -2.552 | -4.373 | -2.202 | -1.944 | 0.498 | 0.803 | 1.401 | 1.417 | -1.423 | -2.202 | -1.944 | -3.095 |
| 09 | -1.738 | 0.301 | -0.149 | -0.626 | -0.511 | 0.276 | -1.082 | -0.626 | -0.188 | 1.001 | 0.555 | -0.756 | -1.423 | 0.325 | 0.803 | -0.407 |
| 10 | 0.325 | -2.202 | -1.423 | -2.202 | 1.822 | -1.568 | -2.202 | -1.296 | 1.508 | -2.202 | -0.828 | -2.202 | 0.574 | -2.552 | -1.296 | -2.552 |
| 11 | 2.189 | -1.082 | 1.335 | -0.228 | -1.296 | -4.373 | -2.202 | -2.202 | -0.359 | -2.552 | -1.423 | -3.095 | -1.568 | -2.552 | -1.568 | -2.552 |
| 12 | 2.151 | -4.373 | 0.348 | -2.552 | -0.906 | -4.373 | -2.552 | -4.373 | 1.197 | -3.095 | -0.008 | -4.373 | -0.626 | -2.202 | -1.082 | -3.095 |
| 13 | 0.226 | -4.373 | 2.434 | -1.738 | -4.373 | -4.373 | -2.202 | -3.095 | -0.689 | -3.095 | 0.803 | -3.095 | -3.095 | -4.373 | -2.552 | -4.373 |
| 14 | -2.552 | -3.095 | 0.457 | -2.202 | -4.373 | -4.373 | -3.095 | -4.373 | 0.024 | -1.296 | 2.357 | 0.679 | -4.373 | -4.373 | -1.423 | -4.373 |
| 15 | -2.202 | -3.095 | -0.27 | -1.568 | -2.552 | -3.095 | -2.202 | -2.202 | -0.567 | 0.348 | 1.479 | 1.8 | -2.552 | -3.095 | -0.906 | 0.457 |
| 16 | -2.552 | -0.99 | -1.296 | -2.202 | -0.828 | -0.567 | -2.552 | -0.756 | 0.226 | 0.773 | 0.172 | 0.056 | -0.27 | 1.448 | 0.695 | -0.076 |
| 17 | 0.477 | -1.568 | -0.828 | -1.296 | 1.704 | -0.99 | -0.359 | -0.228 | 0.887 | -1.568 | -0.689 | -0.906 | 0.348 | -1.296 | -0.458 | -1.423 |
| 18 | 1.156 | 0.914 | 1.264 | 0.555 | -1.568 | -1.183 | -2.202 | -0.828 | -0.689 | -0.314 | -0.567 | -0.756 | -1.183 | -1.183 | -0.906 | -0.359 |