Model info
| Transcription factor | Prdm16 | ||||||||
| Model | PRD16_MOUSE.H11MO.0.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 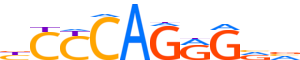 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 10 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bCCCAGRGvv | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.878 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | Factors with multiple dispersed zinc fingers {2.3.4} | ||||||||
| TF subfamily | Evi-1-like factors {2.3.4.14} | ||||||||
| MGI | MGI:1917923 | ||||||||
| EntrezGene | GeneID:70673 (SSTAR profile) | ||||||||
| UniProt ID | PRD16_MOUSE | ||||||||
| UniProt AC | A2A935 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Prdm16 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 34.0 | 249.0 | 64.0 | 153.0 |
| 02 | 13.0 | 410.0 | 8.0 | 69.0 |
| 03 | 27.0 | 392.0 | 4.0 | 77.0 |
| 04 | 28.0 | 462.0 | 1.0 | 9.0 |
| 05 | 494.0 | 5.0 | 1.0 | 0.0 |
| 06 | 43.0 | 4.0 | 434.0 | 19.0 |
| 07 | 133.0 | 7.0 | 327.0 | 33.0 |
| 08 | 78.0 | 9.0 | 410.0 | 3.0 |
| 09 | 158.0 | 90.0 | 234.0 | 18.0 |
| 10 | 246.0 | 118.0 | 101.0 | 35.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.27 | 0.683 | -0.658 | 0.2 |
| 02 | -2.163 | 1.179 | -2.584 | -0.584 |
| 03 | -1.489 | 1.135 | -3.126 | -0.477 |
| 04 | -1.454 | 1.298 | -3.903 | -2.484 |
| 05 | 1.365 | -2.961 | -3.903 | -4.4 |
| 06 | -1.044 | -3.126 | 1.236 | -1.818 |
| 07 | 0.061 | -2.694 | 0.954 | -1.298 |
| 08 | -0.464 | -2.484 | 1.179 | -3.325 |
| 09 | 0.232 | -0.324 | 0.621 | -1.868 |
| 10 | 0.671 | -0.057 | -0.21 | -1.242 |