Model info
| Transcription factor | Pgr | ||||||||
| Model | PRGR_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 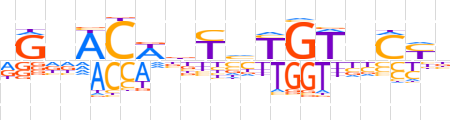 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nRGnACRnRvWGTnCh | ||||||||
| Best auROC (human) | 0.964 | ||||||||
| Best auROC (mouse) | 0.985 | ||||||||
| Peak sets in benchmark (human) | 51 | ||||||||
| Peak sets in benchmark (mouse) | 15 | ||||||||
| Aligned words | 405 | ||||||||
| TF family | Steroid hormone receptors (NR3) {2.1.1} | ||||||||
| TF subfamily | GR-like receptors (NR3C) {2.1.1.1} | ||||||||
| MGI | MGI:97567 | ||||||||
| EntrezGene | |||||||||
| UniProt ID | PRGR_MOUSE | ||||||||
| UniProt AC | Q00175 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Pgr expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 49.0 | 2.0 | 77.0 | 8.0 | 76.0 | 1.0 | 9.0 | 15.0 | 38.0 | 2.0 | 47.0 | 4.0 | 31.0 | 1.0 | 35.0 | 10.0 |
| 02 | 19.0 | 7.0 | 157.0 | 11.0 | 1.0 | 0.0 | 3.0 | 2.0 | 11.0 | 3.0 | 137.0 | 17.0 | 1.0 | 2.0 | 29.0 | 5.0 |
| 03 | 10.0 | 2.0 | 13.0 | 7.0 | 3.0 | 4.0 | 0.0 | 5.0 | 124.0 | 47.0 | 108.0 | 47.0 | 8.0 | 13.0 | 2.0 | 12.0 |
| 04 | 127.0 | 3.0 | 8.0 | 7.0 | 59.0 | 2.0 | 0.0 | 5.0 | 97.0 | 8.0 | 5.0 | 13.0 | 53.0 | 5.0 | 5.0 | 8.0 |
| 05 | 3.0 | 306.0 | 17.0 | 10.0 | 2.0 | 12.0 | 2.0 | 2.0 | 0.0 | 16.0 | 1.0 | 1.0 | 0.0 | 29.0 | 1.0 | 3.0 |
| 06 | 4.0 | 0.0 | 0.0 | 1.0 | 296.0 | 11.0 | 20.0 | 36.0 | 8.0 | 0.0 | 13.0 | 0.0 | 5.0 | 1.0 | 9.0 | 1.0 |
| 07 | 59.0 | 54.0 | 158.0 | 42.0 | 2.0 | 4.0 | 0.0 | 6.0 | 5.0 | 4.0 | 17.0 | 16.0 | 2.0 | 6.0 | 22.0 | 8.0 |
| 08 | 37.0 | 0.0 | 31.0 | 0.0 | 58.0 | 1.0 | 9.0 | 0.0 | 117.0 | 1.0 | 78.0 | 1.0 | 32.0 | 0.0 | 40.0 | 0.0 |
| 09 | 88.0 | 72.0 | 61.0 | 23.0 | 1.0 | 0.0 | 1.0 | 0.0 | 54.0 | 52.0 | 34.0 | 18.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 10 | 23.0 | 31.0 | 8.0 | 82.0 | 33.0 | 10.0 | 1.0 | 80.0 | 28.0 | 10.0 | 1.0 | 57.0 | 4.0 | 4.0 | 1.0 | 32.0 |
| 11 | 0.0 | 4.0 | 84.0 | 0.0 | 19.0 | 4.0 | 31.0 | 1.0 | 0.0 | 2.0 | 8.0 | 1.0 | 8.0 | 3.0 | 240.0 | 0.0 |
| 12 | 3.0 | 2.0 | 5.0 | 17.0 | 3.0 | 2.0 | 0.0 | 8.0 | 30.0 | 13.0 | 16.0 | 304.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 13 | 11.0 | 15.0 | 3.0 | 7.0 | 5.0 | 6.0 | 2.0 | 4.0 | 6.0 | 8.0 | 3.0 | 4.0 | 66.0 | 101.0 | 58.0 | 106.0 |
| 14 | 7.0 | 68.0 | 1.0 | 12.0 | 15.0 | 95.0 | 1.0 | 19.0 | 8.0 | 51.0 | 3.0 | 4.0 | 4.0 | 108.0 | 2.0 | 7.0 |
| 15 | 6.0 | 14.0 | 10.0 | 4.0 | 50.0 | 113.0 | 6.0 | 153.0 | 0.0 | 2.0 | 4.0 | 1.0 | 1.0 | 14.0 | 8.0 | 19.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.653 | -2.381 | 1.103 | -1.121 | 1.09 | -2.927 | -1.008 | -0.513 | 0.401 | -2.381 | 0.612 | -1.77 | 0.2 | -2.927 | 0.32 | -0.907 |
| 02 | -0.282 | -1.248 | 1.813 | -0.815 | -2.927 | -4.226 | -2.03 | -2.381 | -0.815 | -2.03 | 1.677 | -0.391 | -2.927 | -2.381 | 0.134 | -1.564 |
| 03 | -0.907 | -2.381 | -0.653 | -1.248 | -2.03 | -1.77 | -4.226 | -1.564 | 1.577 | 0.612 | 1.44 | 0.612 | -1.121 | -0.653 | -2.381 | -0.73 |
| 04 | 1.601 | -2.03 | -1.121 | -1.248 | 0.838 | -2.381 | -4.226 | -1.564 | 1.333 | -1.121 | -1.564 | -0.653 | 0.731 | -1.564 | -1.564 | -1.121 |
| 05 | -2.03 | 2.479 | -0.391 | -0.907 | -2.381 | -0.73 | -2.381 | -2.381 | -4.226 | -0.45 | -2.927 | -2.927 | -4.226 | 0.134 | -2.927 | -2.03 |
| 06 | -1.77 | -4.226 | -4.226 | -2.927 | 2.446 | -0.815 | -0.232 | 0.348 | -1.121 | -4.226 | -0.653 | -4.226 | -1.564 | -2.927 | -1.008 | -2.927 |
| 07 | 0.838 | 0.75 | 1.819 | 0.501 | -2.381 | -1.77 | -4.226 | -1.394 | -1.564 | -1.77 | -0.391 | -0.45 | -2.381 | -1.394 | -0.138 | -1.121 |
| 08 | 0.375 | -4.226 | 0.2 | -4.226 | 0.821 | -2.927 | -1.008 | -4.226 | 1.519 | -2.927 | 1.115 | -2.927 | 0.231 | -4.226 | 0.452 | -4.226 |
| 09 | 1.236 | 1.036 | 0.871 | -0.094 | -2.927 | -4.226 | -2.927 | -4.226 | 0.75 | 0.712 | 0.291 | -0.335 | -2.927 | -4.226 | -4.226 | -4.226 |
| 10 | -0.094 | 0.2 | -1.121 | 1.165 | 0.262 | -0.907 | -2.927 | 1.141 | 0.1 | -0.907 | -2.927 | 0.804 | -1.77 | -1.77 | -2.927 | 0.231 |
| 11 | -4.226 | -1.77 | 1.189 | -4.226 | -0.282 | -1.77 | 0.2 | -2.927 | -4.226 | -2.381 | -1.121 | -2.927 | -1.121 | -2.03 | 2.236 | -4.226 |
| 12 | -2.03 | -2.381 | -1.564 | -0.391 | -2.03 | -2.381 | -4.226 | -1.121 | 0.168 | -0.653 | -0.45 | 2.472 | -4.226 | -4.226 | -4.226 | -2.381 |
| 13 | -0.815 | -0.513 | -2.03 | -1.248 | -1.564 | -1.394 | -2.381 | -1.77 | -1.394 | -1.121 | -2.03 | -1.77 | 0.949 | 1.373 | 0.821 | 1.421 |
| 14 | -1.248 | 0.979 | -2.927 | -0.73 | -0.513 | 1.312 | -2.927 | -0.282 | -1.121 | 0.693 | -2.03 | -1.77 | -1.77 | 1.44 | -2.381 | -1.248 |
| 15 | -1.394 | -0.581 | -0.907 | -1.77 | 0.673 | 1.485 | -1.394 | 1.787 | -4.226 | -2.381 | -1.77 | -2.927 | -2.927 | -0.581 | -1.121 | -0.282 |