Model info
| Transcription factor | Pgr | ||||||||
| Model | PRGR_MOUSE.H11MO.1.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 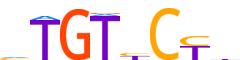 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 8 | ||||||||
| Quality | A | ||||||||
| Motif rank | 1 | ||||||||
| Consensus | vRGdACAb | ||||||||
| Best auROC (human) | 0.845 | ||||||||
| Best auROC (mouse) | 0.924 | ||||||||
| Peak sets in benchmark (human) | 51 | ||||||||
| Peak sets in benchmark (mouse) | 15 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Steroid hormone receptors (NR3) {2.1.1} | ||||||||
| TF subfamily | GR-like receptors (NR3C) {2.1.1.1} | ||||||||
| MGI | MGI:97567 | ||||||||
| EntrezGene | |||||||||
| UniProt ID | PRGR_MOUSE | ||||||||
| UniProt AC | Q00175 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Pgr expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 186.0 | 111.0 | 144.0 | 59.0 |
| 02 | 279.0 | 1.0 | 202.0 | 18.0 |
| 03 | 9.0 | 1.0 | 486.0 | 4.0 |
| 04 | 242.0 | 49.0 | 153.0 | 56.0 |
| 05 | 467.0 | 11.0 | 6.0 | 16.0 |
| 06 | 5.0 | 484.0 | 9.0 | 2.0 |
| 07 | 452.0 | 3.0 | 5.0 | 40.0 |
| 08 | 64.0 | 97.0 | 214.0 | 125.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.393 | -0.117 | 0.14 | -0.737 |
| 02 | 0.796 | -3.903 | 0.475 | -1.868 |
| 03 | -2.484 | -3.903 | 1.349 | -3.126 |
| 04 | 0.655 | -0.918 | 0.2 | -0.788 |
| 05 | 1.309 | -2.311 | -2.819 | -1.975 |
| 06 | -2.961 | 1.345 | -2.484 | -3.573 |
| 07 | 1.276 | -3.325 | -2.961 | -1.114 |
| 08 | -0.658 | -0.25 | 0.533 | 0.0 |