Model info
| Transcription factor | RARA (GeneCards) | ||||||||
| Model | RARA_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 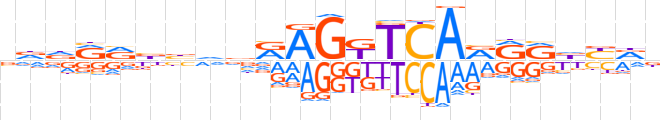 | ||||||||
| LOGO (reverse complement) | 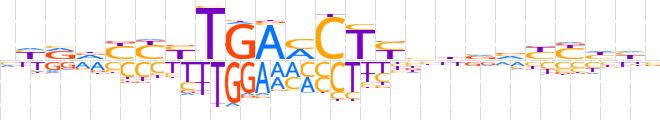 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vddRRbbvvRAGKTCARRRbYRn | ||||||||
| Best auROC (human) | 0.954 | ||||||||
| Best auROC (mouse) | 0.935 | ||||||||
| Peak sets in benchmark (human) | 29 | ||||||||
| Peak sets in benchmark (mouse) | 32 | ||||||||
| Aligned words | 356 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | Retinoic acid receptors (NR1B) {2.1.2.1} | ||||||||
| HGNC | HGNC:9864 | ||||||||
| EntrezGene | GeneID:5914 (SSTAR profile) | ||||||||
| UniProt ID | RARA_HUMAN | ||||||||
| UniProt AC | P10276 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | RARA expression | ||||||||
| ReMap ChIP-seq dataset list | RARA datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 37.0 | 7.0 | 29.0 | 7.0 | 44.0 | 9.0 | 5.0 | 23.0 | 80.0 | 8.0 | 50.0 | 13.0 | 11.0 | 3.0 | 15.0 | 6.0 |
| 02 | 55.0 | 7.0 | 96.0 | 14.0 | 7.0 | 5.0 | 5.0 | 10.0 | 30.0 | 4.0 | 46.0 | 19.0 | 12.0 | 6.0 | 18.0 | 13.0 |
| 03 | 10.0 | 6.0 | 82.0 | 6.0 | 6.0 | 6.0 | 4.0 | 6.0 | 27.0 | 7.0 | 125.0 | 6.0 | 12.0 | 24.0 | 14.0 | 6.0 |
| 04 | 18.0 | 7.0 | 23.0 | 7.0 | 30.0 | 8.0 | 0.0 | 5.0 | 67.0 | 7.0 | 142.0 | 9.0 | 8.0 | 5.0 | 11.0 | 0.0 |
| 05 | 13.0 | 23.0 | 45.0 | 42.0 | 11.0 | 7.0 | 1.0 | 8.0 | 14.0 | 27.0 | 30.0 | 105.0 | 1.0 | 2.0 | 3.0 | 15.0 |
| 06 | 7.0 | 9.0 | 10.0 | 13.0 | 24.0 | 17.0 | 3.0 | 15.0 | 6.0 | 22.0 | 16.0 | 35.0 | 7.0 | 118.0 | 31.0 | 14.0 |
| 07 | 3.0 | 4.0 | 30.0 | 7.0 | 127.0 | 20.0 | 5.0 | 14.0 | 19.0 | 10.0 | 23.0 | 8.0 | 11.0 | 25.0 | 13.0 | 28.0 |
| 08 | 46.0 | 14.0 | 89.0 | 11.0 | 32.0 | 10.0 | 1.0 | 16.0 | 26.0 | 5.0 | 25.0 | 15.0 | 4.0 | 29.0 | 12.0 | 12.0 |
| 09 | 54.0 | 5.0 | 45.0 | 4.0 | 48.0 | 2.0 | 6.0 | 2.0 | 63.0 | 4.0 | 60.0 | 0.0 | 11.0 | 8.0 | 32.0 | 3.0 |
| 10 | 137.0 | 1.0 | 37.0 | 1.0 | 17.0 | 1.0 | 1.0 | 0.0 | 117.0 | 0.0 | 23.0 | 3.0 | 7.0 | 0.0 | 2.0 | 0.0 |
| 11 | 17.0 | 2.0 | 257.0 | 2.0 | 0.0 | 0.0 | 2.0 | 0.0 | 3.0 | 1.0 | 57.0 | 2.0 | 1.0 | 0.0 | 3.0 | 0.0 |
| 12 | 2.0 | 0.0 | 6.0 | 13.0 | 0.0 | 3.0 | 0.0 | 0.0 | 6.0 | 6.0 | 154.0 | 153.0 | 1.0 | 0.0 | 1.0 | 2.0 |
| 13 | 0.0 | 0.0 | 3.0 | 6.0 | 2.0 | 2.0 | 0.0 | 5.0 | 5.0 | 13.0 | 14.0 | 129.0 | 2.0 | 0.0 | 2.0 | 164.0 |
| 14 | 0.0 | 8.0 | 1.0 | 0.0 | 5.0 | 9.0 | 0.0 | 1.0 | 1.0 | 17.0 | 1.0 | 0.0 | 5.0 | 272.0 | 7.0 | 20.0 |
| 15 | 5.0 | 0.0 | 5.0 | 1.0 | 301.0 | 0.0 | 4.0 | 1.0 | 9.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 | 1.0 | 0.0 |
| 16 | 204.0 | 28.0 | 81.0 | 22.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 2.0 | 3.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 17 | 21.0 | 7.0 | 175.0 | 6.0 | 8.0 | 1.0 | 14.0 | 7.0 | 38.0 | 5.0 | 39.0 | 4.0 | 1.0 | 11.0 | 8.0 | 2.0 |
| 18 | 33.0 | 10.0 | 23.0 | 2.0 | 3.0 | 14.0 | 0.0 | 7.0 | 18.0 | 7.0 | 205.0 | 6.0 | 1.0 | 9.0 | 6.0 | 3.0 |
| 19 | 15.0 | 3.0 | 30.0 | 7.0 | 13.0 | 7.0 | 1.0 | 19.0 | 22.0 | 38.0 | 19.0 | 155.0 | 1.0 | 7.0 | 4.0 | 6.0 |
| 20 | 4.0 | 19.0 | 15.0 | 13.0 | 7.0 | 36.0 | 1.0 | 11.0 | 3.0 | 6.0 | 13.0 | 32.0 | 5.0 | 141.0 | 23.0 | 18.0 |
| 21 | 4.0 | 1.0 | 13.0 | 1.0 | 167.0 | 14.0 | 8.0 | 13.0 | 14.0 | 3.0 | 21.0 | 14.0 | 21.0 | 9.0 | 22.0 | 22.0 |
| 22 | 25.0 | 33.0 | 87.0 | 61.0 | 16.0 | 6.0 | 2.0 | 3.0 | 7.0 | 9.0 | 28.0 | 20.0 | 3.0 | 33.0 | 8.0 | 6.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.527 | -1.097 | 0.286 | -1.097 | 0.699 | -0.856 | -1.413 | 0.058 | 1.293 | -0.969 | 0.826 | -0.501 | -0.663 | -1.88 | -0.361 | -1.243 |
| 02 | 0.921 | -1.097 | 1.475 | -0.429 | -1.097 | -1.413 | -1.413 | -0.755 | 0.32 | -1.62 | 0.743 | -0.13 | -0.579 | -1.243 | -0.183 | -0.501 |
| 03 | -0.755 | -1.243 | 1.318 | -1.243 | -1.243 | -1.243 | -1.62 | -1.243 | 0.216 | -1.097 | 1.738 | -1.243 | -0.579 | 0.1 | -0.429 | -1.243 |
| 04 | -0.183 | -1.097 | 0.058 | -1.097 | 0.32 | -0.969 | -4.1 | -1.413 | 1.117 | -1.097 | 1.865 | -0.856 | -0.969 | -1.413 | -0.663 | -4.1 |
| 05 | -0.501 | 0.058 | 0.721 | 0.653 | -0.663 | -1.097 | -2.782 | -0.969 | -0.429 | 0.216 | 0.32 | 1.564 | -2.782 | -2.232 | -1.88 | -0.361 |
| 06 | -1.097 | -0.856 | -0.755 | -0.501 | 0.1 | -0.239 | -1.88 | -0.361 | -1.243 | 0.014 | -0.298 | 0.472 | -1.097 | 1.68 | 0.352 | -0.429 |
| 07 | -1.88 | -1.62 | 0.32 | -1.097 | 1.754 | -0.08 | -1.413 | -0.429 | -0.13 | -0.755 | 0.058 | -0.969 | -0.663 | 0.14 | -0.501 | 0.252 |
| 08 | 0.743 | -0.429 | 1.399 | -0.663 | 0.384 | -0.755 | -2.782 | -0.298 | 0.179 | -1.413 | 0.14 | -0.361 | -1.62 | 0.286 | -0.579 | -0.579 |
| 09 | 0.902 | -1.413 | 0.721 | -1.62 | 0.785 | -2.232 | -1.243 | -2.232 | 1.055 | -1.62 | 1.007 | -4.1 | -0.663 | -0.969 | 0.384 | -1.88 |
| 10 | 1.829 | -2.782 | 0.527 | -2.782 | -0.239 | -2.782 | -2.782 | -4.1 | 1.672 | -4.1 | 0.058 | -1.88 | -1.097 | -4.1 | -2.232 | -4.1 |
| 11 | -0.239 | -2.232 | 2.457 | -2.232 | -4.1 | -4.1 | -2.232 | -4.1 | -1.88 | -2.782 | 0.956 | -2.232 | -2.782 | -4.1 | -1.88 | -4.1 |
| 12 | -2.232 | -4.1 | -1.243 | -0.501 | -4.1 | -1.88 | -4.1 | -4.1 | -1.243 | -1.243 | 1.946 | 1.939 | -2.782 | -4.1 | -2.782 | -2.232 |
| 13 | -4.1 | -4.1 | -1.88 | -1.243 | -2.232 | -2.232 | -4.1 | -1.413 | -1.413 | -0.501 | -0.429 | 1.769 | -2.232 | -4.1 | -2.232 | 2.009 |
| 14 | -4.1 | -0.969 | -2.782 | -4.1 | -1.413 | -0.856 | -4.1 | -2.782 | -2.782 | -0.239 | -2.782 | -4.1 | -1.413 | 2.514 | -1.097 | -0.08 |
| 15 | -1.413 | -4.1 | -1.413 | -2.782 | 2.615 | -4.1 | -1.62 | -2.782 | -0.856 | -4.1 | -4.1 | -4.1 | -0.08 | -4.1 | -2.782 | -4.1 |
| 16 | 2.226 | 0.252 | 1.305 | 0.014 | -4.1 | -4.1 | -4.1 | -4.1 | -1.413 | -2.232 | -1.88 | -4.1 | -4.1 | -4.1 | -2.232 | -4.1 |
| 17 | -0.032 | -1.097 | 2.073 | -1.243 | -0.969 | -2.782 | -0.429 | -1.097 | 0.554 | -1.413 | 0.579 | -1.62 | -2.782 | -0.663 | -0.969 | -2.232 |
| 18 | 0.414 | -0.755 | 0.058 | -2.232 | -1.88 | -0.429 | -4.1 | -1.097 | -0.183 | -1.097 | 2.231 | -1.243 | -2.782 | -0.856 | -1.243 | -1.88 |
| 19 | -0.361 | -1.88 | 0.32 | -1.097 | -0.501 | -1.097 | -2.782 | -0.13 | 0.014 | 0.554 | -0.13 | 1.952 | -2.782 | -1.097 | -1.62 | -1.243 |
| 20 | -1.62 | -0.13 | -0.361 | -0.501 | -1.097 | 0.5 | -2.782 | -0.663 | -1.88 | -1.243 | -0.501 | 0.384 | -1.413 | 1.858 | 0.058 | -0.183 |
| 21 | -1.62 | -2.782 | -0.501 | -2.782 | 2.027 | -0.429 | -0.969 | -0.501 | -0.429 | -1.88 | -0.032 | -0.429 | -0.032 | -0.856 | 0.014 | 0.014 |
| 22 | 0.14 | 0.414 | 1.377 | 1.023 | -0.298 | -1.243 | -2.232 | -1.88 | -1.097 | -0.856 | 0.252 | -0.08 | -1.88 | 0.414 | -0.969 | -1.243 |